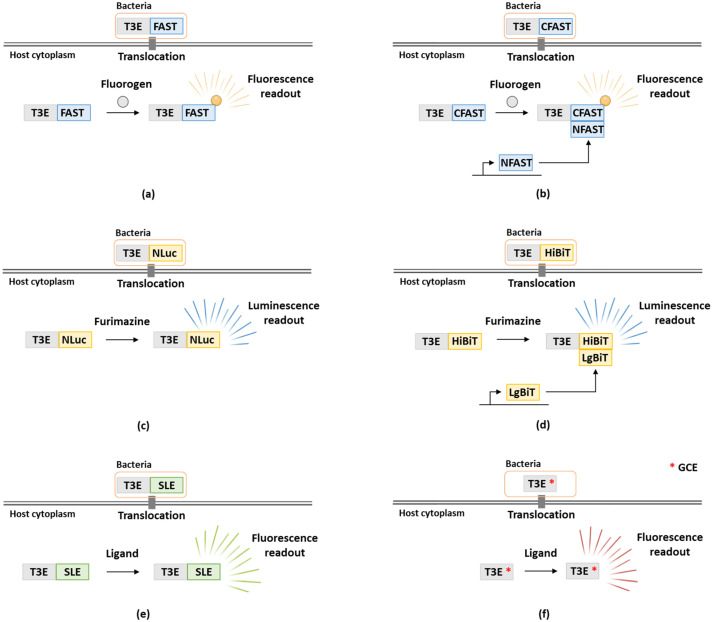
Figure 1.
Expansion to the toolset to study bacterial effector translocation. (a) The FAST-based method was recently developed to monitor bacterial effector translocation. To this end, bacterial effectors are fused to the FAST tag. Upon translocation and addition of fluorogen, fluorescence can be measured. (b) Hypothetically, a split-FAST system can also be used. Here, the shorter C-terminal fragment of FAST (CFAST) is fused to the effector of interest, whilst the complementing N-terminal fragment (NFAST) is expressed inside host cells. Upon translocation, complementation takes place, resulting in fluorescence in the presence of fluorogen. (c) The use of NanoLuc (NLuc) to study bacterial effector translocation. Upon translocation of the effector fused to NLuc and addition of furimazine, luminescence is measured. (d) Similar to split-FAST, a split-NanoLuc complementation-based system (NanoBiT) allows real-time monitoring of effector translocation. In this case, HiBiT is fused to the effector of interest and LgBiT is expressed inside host cells. (e) The use of self-labeling enzyme (SLE) tags (e.g., HaloTag, CLIP or SNAP) to monitor effector translocation upon addition of the appropriate ligand. (f) Using genetic code expansion (GCE), a non-canonical amino acid (indicated with a red *) is incorporated into the effector under study, allowing monitoring of translocation inside host cells.