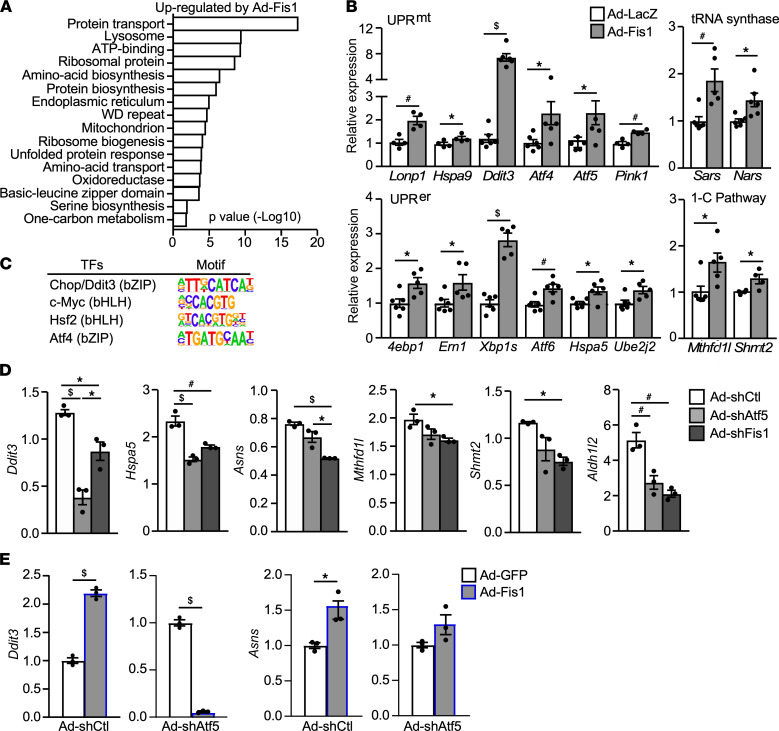
Figure 3. Fis1 overexpression induces an ISR.
(A) Functional clustering analysis of 1236 genes upregulated (FDR P < 0.001) by Ad-Fis1 versus Ad-LacZ in primary hepatocytes identified by RNA-Seq. n = 4 in 1 experiment. (B) Validation of RNA-Seq results for ISR genes in UPRmt, UPRer, tRNA synthesis, and 1-carbon metabolism (1-C) pathway in primary hepatocytes infected with Ad-Fis1 or Ad-LacZ by real-time qPCR. n = 4–6. (C) HOMER motif analysis to identify potential transcription factor binding sites on promoters of Ad-Fis1–upregulated genes. (D) Assessing the effects of knocking down Atf5 or Fis1 on ISR gene expression in hepatocytes by real-time PCR analysis. Primary hepatocytes were infected with Ad-shCtl, Ad-shAtf5, or Ad-shFis1 and cells were harvested 48 hours after infection. 36b4 was used for normalization to determine the relative expression. n = 3, repeated 4 times. (E) Assessing Atf5 as a Fis1 downstream effector in regulating hepatic ISR gene expression using real-time PCR. Primary hepatocytes were infected with Ad-shCtl or Ad-shAtf5; 8 hours later, cells were washed and infected with Ad-GFP or Ad-Fis1. Hepatocytes were cultured for an additional 40 hours. n = 3, repeated twice. Values are presented as mean ± SEM. Significance of B and E were determined by unpaired, 2-tailed Student’s t test; and of D by 1-way ANOVA followed by Holm-Šidák multiple comparisons test. *P < 0.05; #P < 0.01; $P < 0.001.