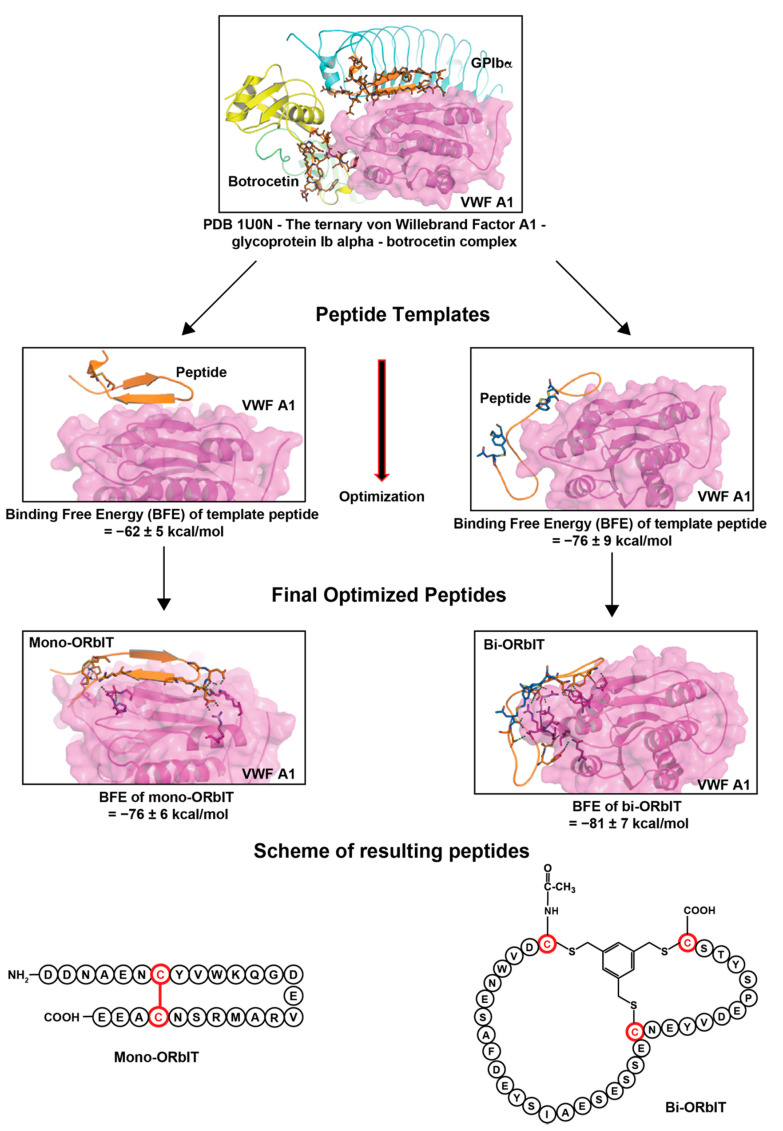
Figure 1.
Design of GPIbα-mimicking peptides to interfere with VWF A1 domain. Molecular structures of the ternary complexes of GPIbα-VWF A1 and GPIbα-VWF A1–botrocetin (PDB code: 1U0N) were subjected to molecular dynamics simulations. Low energy structures shown of GPIbα (blue), botrocetin (yellow), and VWF A1 domain (purple). The indicated residues of GPIbα with (out) botrocetin identified as interacting with VWF A1 were used for the in silico design of (>100) mono and bi-cyclic peptides. All designed peptides were subjected to molecular dynamics simulations and binding free-energy (BFE) calculations. Peptide mutants with lowest BFE were selected for optimization and synthesis: mono-ORbIT (−76 ± 6 kcal/mol) and bi-ORbIT (−81 ± 7 kcal/mol).