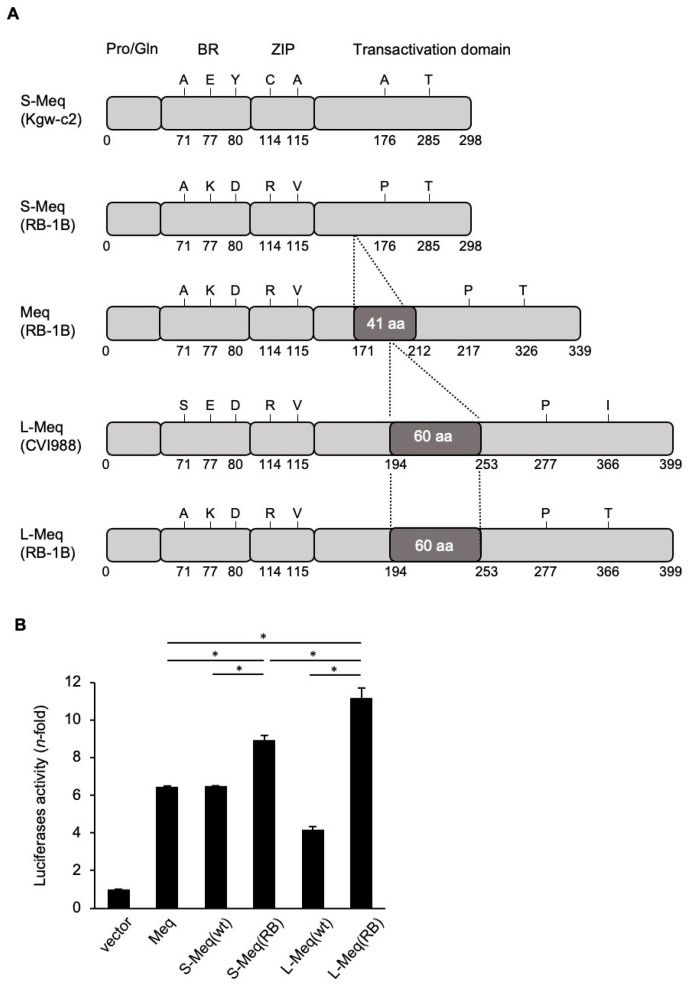
Figure 1.
Analysis of the transactivation activity of the three Meq isoforms. (A) Schematic representation of the different Meq isoforms. The structures of short-Meq containing the deletion (S-Meq) from Kgw-c2, a Marek’s disease virus (MDV) strain, Meq from RB-1B, and long-Meq containing the insertion (L-Meq) from CVI988, and S-Meq (RB-1B) and L-Meq (RB-1B) whose sequences were matched with that of RB-1B-Meq, except for the deletion or insertion in the transactivation domain, are indicated. Meq comprises a proline/glutamine (Pro/Gln) rich domain followed by the basic region (BR) and leucine zipper (ZIP) at the N-terminal region, and the transactivation domain at the C-terminal region. The Meq isoforms include mutations in the BR, ZIP, and transactivation domain. S-Meq contains a 41 amino acid deletion in the transactivation domain and L-Meq is characterized by a 60-amino acid insertion in the transactivation domain. (B) Transactivation activity of the Meq isoforms. The transactivation activity of RB-1B-Meq, wild-type S-Meq (Kgw-c2), wild-type L-Meq (CVI988), S-Meq (RB-1B), and L-Meq (RB-1B) was compared on the Meq promoter-driven luciferase activities. DF-1 cells in each well were transfected with 300 ng of expression plasmids, Meq, S-Meq (wild-type), S-Meq (RB-1B), L-Meq (wild-type), or L-Meq (RB-1B); 200 ng of the c-Jun expression plasmid; 500 ng of the reporter plasmid; and 5 ng of control pRL-TK. Luciferase activities were analyzed 24 h post-transfection. Firefly luciferase activity is expressed relative to the mean basal activity in the presence of pCI-neo after normalization to Renilla luciferase activity. Error bars indicate standard deviations. * p < 0.01.