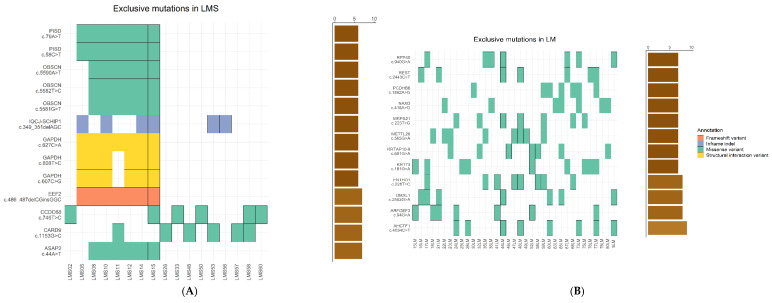
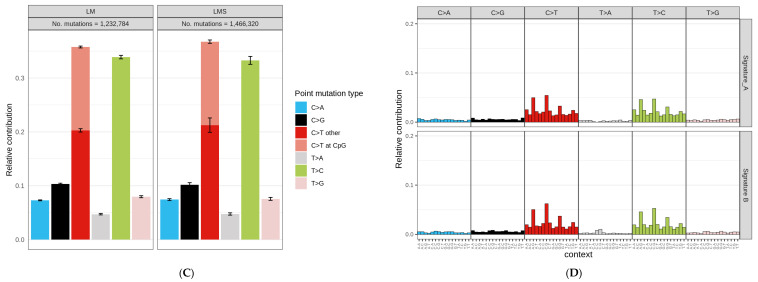
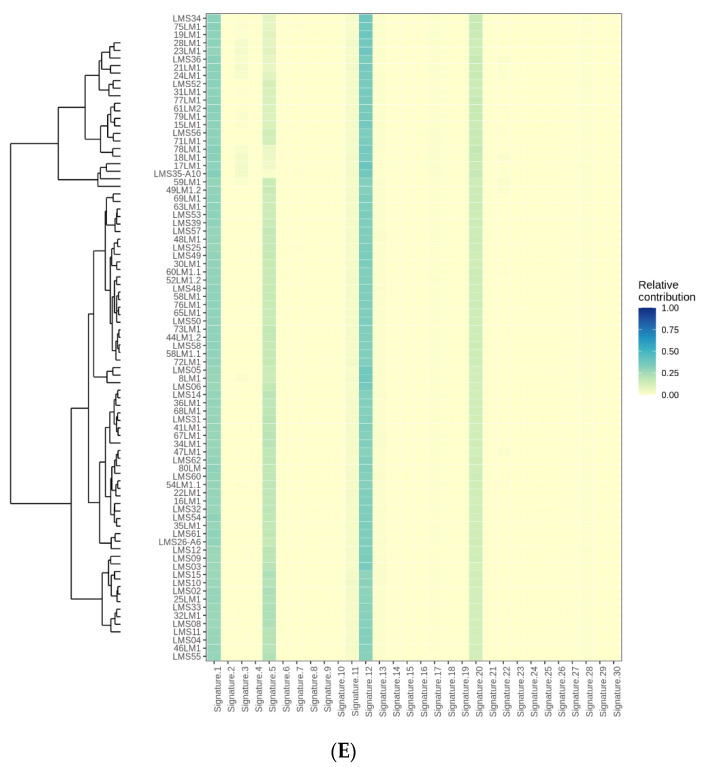
Figure 1.
Comparative analysis of single-nucleotide variants (SNVs), insertions/deletions (indels), and mutational signatures for leiomyosarcoma (LMS) and leiomyoma (LM) samples. (A) Tumor profile of LMS-exclusive variants, including frequency and type of mutations. (B) Tumor profile of LM-exclusive variants, including frequency and type of mutations. In both cases, rows represent individual genes, while columns represent individual tumors. Bars illustrate the number of samples for each exclusive mutation. Types of mutations are annotated according to color. (C) Relative contribution of the indicated mutation types to the point mutation spectrum for each tumor type. Error bars indicate standard deviation over all samples. Total number of mutations for LM and LMS is indicated. (D) Relative contribution of each indicated trinucleotide changes to the two mutational signatures identified by non-negative matrix factorization (NMF) analysis. (E) Heatmap showing relative contribution of each mutational signature described in the COSMIC database for each sample.