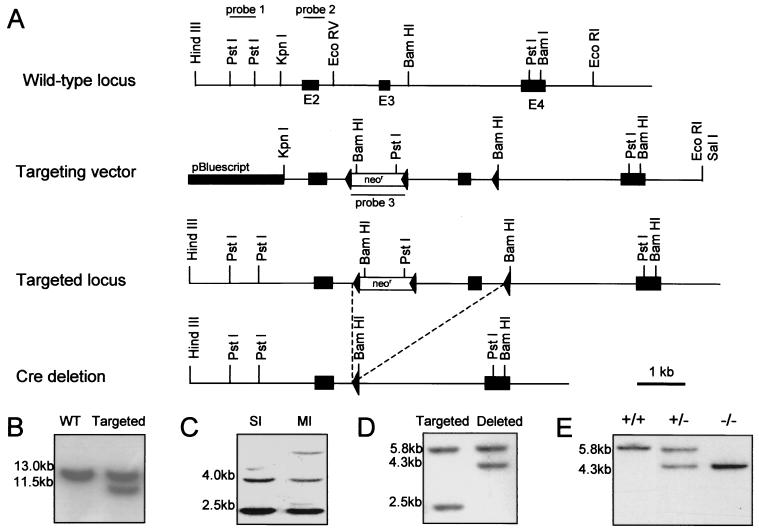
FIG. 1.
Targeted disruption of the NID-1 gene and generation of chimeras. (A) A restriction map and exon-intron structure (top), the targeting vector (middle), and the mutant allele after homologous recombination and Cre recombinase-mediated deletion (bottom) are shown. Exon numbers are indicated. (B) BamHI-digested DNA from ES clones analyzed with probe 1. The wild-type (WT) and targeted alleles generate 13- and 11.5-kb fragments, respectively. (C) PstI-digested DNA from ES cells analyzed with probe 3 revealed two bands of 2.5 and 4 kb for single integration (SI) of the targeting construct and additional bands in the case of multiple integrations (MI). (D) Southern blot of genomic DNA from ES clones after transient transfection with the Cre recombinase expression plasmid analyzed with probe 2. Hybridization of PstI-digested DNA yields for the targeted allele bands of 5.8 and 2.5 kb and, after Cre recombinase-mediated deletion of the neoR cassette and exon 3, a shift of the smaller band to 4.3 kb. (E) BamHI-digested tail DNA of the offspring from NID-1+/− mice matings hybridized with probe 2 show that homozygous animals were born.