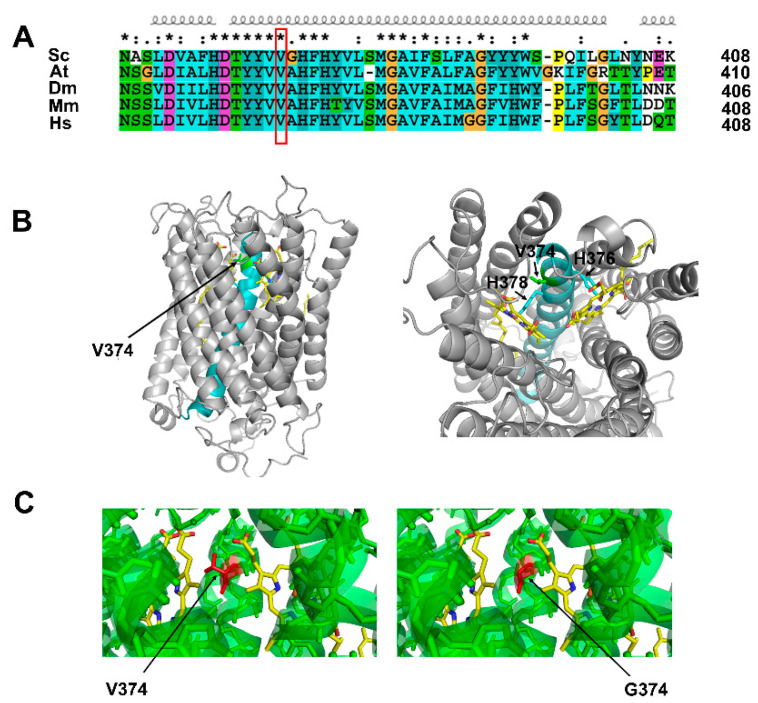
Figure 4.
Cox1 p.Val374Gly mutation did not affect the protein tertiary structure. (A) Alignment of yeast Cox1 sequences containing Val374 in different species. Sequences of Saccharomyces cerevisiae (Sc), Aradopsis thaliana (At), Drosophila melanogaster (Dm), Mus musculus (Mm), and Homo sapiens (Hs) are aligned with ClustalX 2.1. Identical and conserved residues were marked with asterisks. Val374 is marked with a red rectangle, and the α-helices are marked on the top of sequences. (B) Side view (left panel) and top view (right panel) of Cox1 from Saccharomyces cerevisiae (PDB code: 6hu9). The transmembrane α-helix containing Val374 is colored in cyan, Val374 in green, His376 and His378 in blue, and two hemes in yellow. (C) Tertiary structure of wild-type yeast Cox1 (PDB code: 6hu9; left panel) and Cox1 p.Val374Gly mutation (predicted by SWISSMODEL; right panel). The transmembrane α-helix is colored in green, residue 374 in red, and two hemes in yellow.