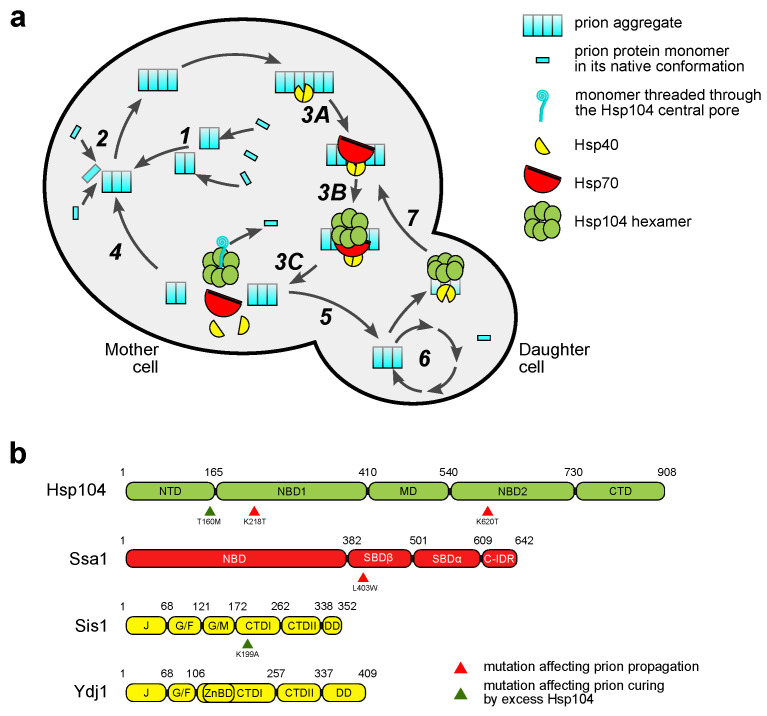
Figure 1.
Control of the yeast prion life cycle by PQC machinery. (a) A schematic representation of the yeast prion life cycle (based on [41] with modifications). 1—de novo aggregate formation; 2—growth of a newly formed aggregate; 3—chaperone-dependent fragmentation, which includes recognition by Hsp40s, followed by recruitment of Hsp70 (3A), interaction with Hsp104 (3B) and aggregate shearing (3C); 4—continuation of the cycle in mother cell; 5—transmission of prion seed into daughter cell; 6—continuation of the cycle in daughter cell; 7—retaining of an aggregate in mother cell by retrograde transport or asymmetric inheritance. (b) The domain structure of the PQC components involved in yeast prion propagation. NTD—N-terminal domain; NBD—nucleotide-binding domain; MD—middle domain; CTD—C-terminal domain; SBD—substrate-binding domain; C-IDR—C-terminal intrinsically disordered region; J—J-domain; G/F—glycine/phenylalanine-rich region; G/M—glycine/methionine-rich region; ZnBD—Zn2+-binding domain or region; DD—dimerization domain. Domain boundaries are taken from [42] for Hsp104, [43] for Ssa1, [44] for Sis1, [45] for Ydj1. Arrowheads point to locations of most notable mutations affecting prion propagation or prion curing by excess Hsp104.