Fig. 2. Expansion and emergence of antibiotic-resistant S. epidermidis within skin-associated microbial communities.
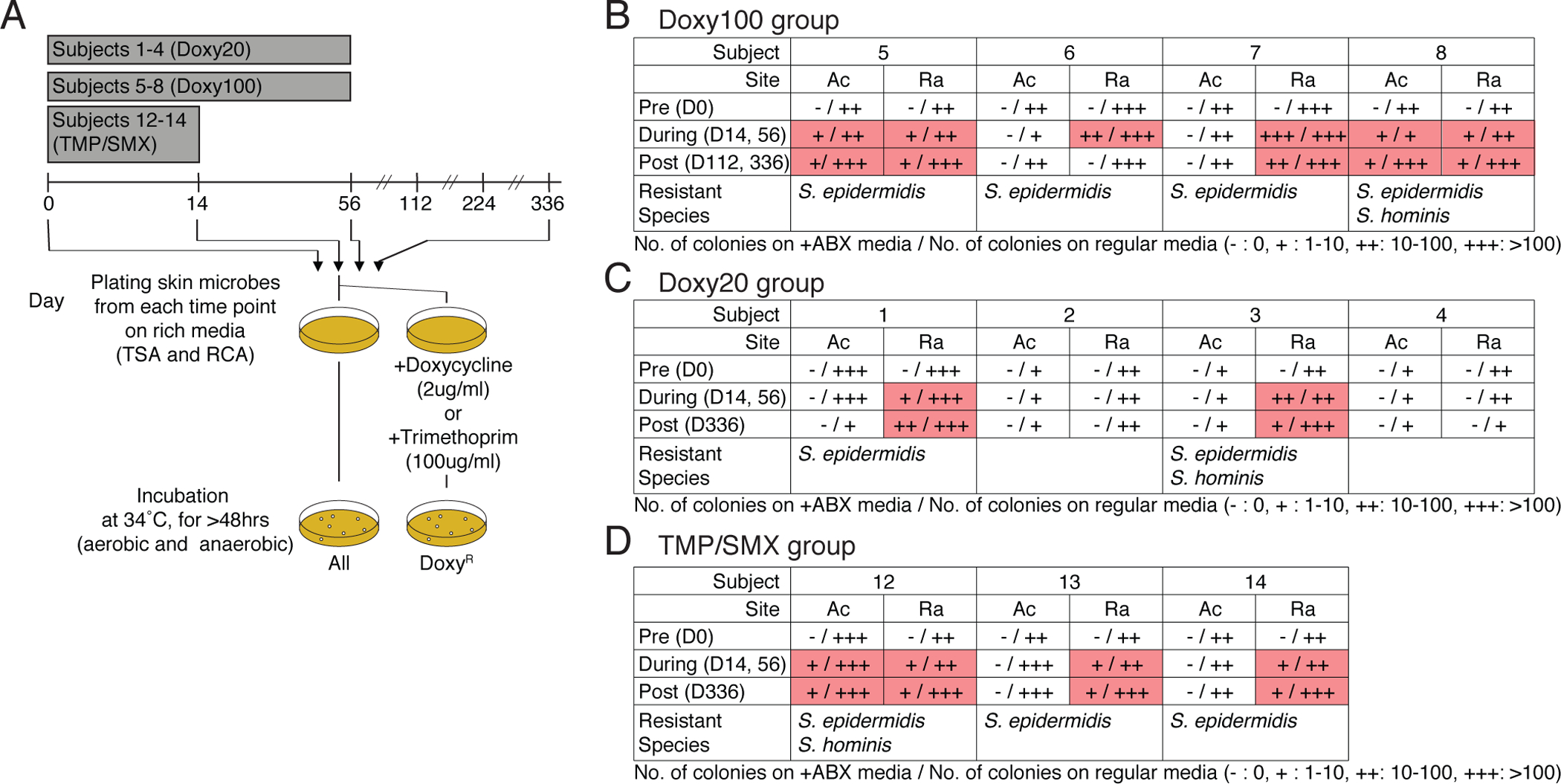
(A) Experimental design for in vitro culturing of doxycycline-resistant (DoxyR) or TMP-resistant (TMPR)skin bacteria. Skin swabs from D0, D14, D56, D112, and D336 were plated on TSB and RCA agar plate (with or without Doxy 2ug/ml or 100ug/ml), and incubated under aerobic and anaerobic conditions, respectively. After 72 hours (up to 120 hours to allow for minor and slow growing microbes to make visible colony), colonies were counted, grown independently, DNA extracted, and sequenced. (B) to (D) Summary tables for in vitro culture results of (B) Doxy100 subjects, (C) Doxy20 subjects and (D) TMP/SMX subjects. (+: 1–10 colonies, ++: 10–100 colonies, +++: >100 colonies, pink boxes highlight presence of DoxyR colonies). Cells with read shading indicate where resistant colonies were found. Species of isolated resistant bactera are listed at the bottom of each table.
