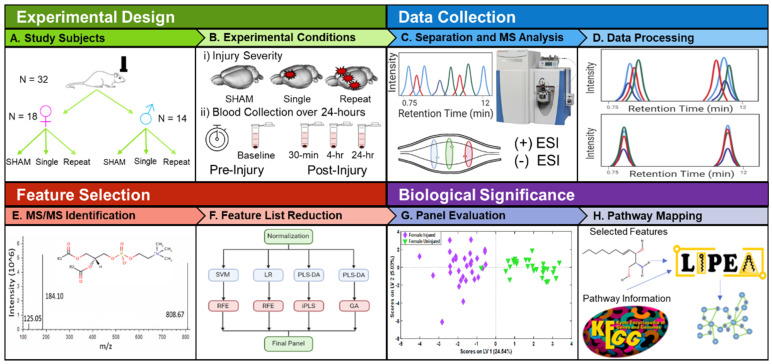
Figure 1.
Overview of study design, data processing, feature selection and identification. (A) Experimental groups included both male (n = 14) and female (n = 18) sexes and were assigned to sham controls that received no injuries (n = 11), single impact that received one closed head impact (n = 10), or repeat impact that received three separate closed head impacts (n = 11). (B) Injury groups and whole blood collection. (C) Workflow illustrating LC-MS data collection in both positive and negative ion modes (D) Peak alignment, picking and integration were accomplished using Compound Discoverer v.3.0, a Thermo Scientific software. (E) Identification of known lipid species using MSMS spectra collected with data-dependent acquisition (DDA) and in-house databases. (F) Multivariate model development and feature selection using machine learning methods to identify features most relevant to differentiating control and TBI classes. (G) Features identified by two or more machine learning approaches were combined to create the final oPLS-DA models. (H) Features selected in the final panels were imported into LIPEA to determine alignment of lipids with biological pathways altered following TBI.