FIG. 2.
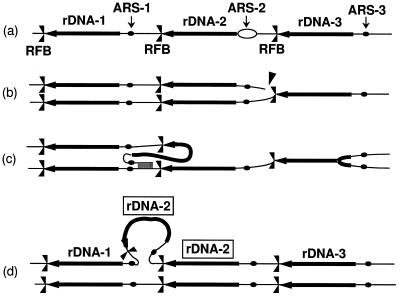
The fork block-dependent recombination model for rDNA repeat expansion and contraction. The positions of ARS and RFB are shown as solid dots and  , respectively. Individual lines represent chromatids with double-stranded DNA. In this model, DNA replication starts from one of the ARS sites (ARS-2) bidirectionally (a). In the yeast rDNA repeats, about one in five ARS sites is used as an active origin (2, 21). A rightward replication fork is arrested at the RFB site, and this arrest is supposed to stimulate a double-strand break of DNA at a nearby site (indicated by an arrowhead in row b). A strand invasion at a homologous duplex (a downstream sister chromatid near ARS-1 in this example) takes place (c), and a new replication fork is formed. The new replication fork meets with the leftward replication fork from the upstream site, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as boxed rDNA-2 (d). If the strand invasion is at a site in a upstream repeat (e.g., near ARS-3), a loss, rather than a gain, of an rDNA repeat is expected. This model was proposed previously to explain the observed strong dependence of rDNA repeat expansion and contraction on FOB1 (18).
, respectively. Individual lines represent chromatids with double-stranded DNA. In this model, DNA replication starts from one of the ARS sites (ARS-2) bidirectionally (a). In the yeast rDNA repeats, about one in five ARS sites is used as an active origin (2, 21). A rightward replication fork is arrested at the RFB site, and this arrest is supposed to stimulate a double-strand break of DNA at a nearby site (indicated by an arrowhead in row b). A strand invasion at a homologous duplex (a downstream sister chromatid near ARS-1 in this example) takes place (c), and a new replication fork is formed. The new replication fork meets with the leftward replication fork from the upstream site, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as boxed rDNA-2 (d). If the strand invasion is at a site in a upstream repeat (e.g., near ARS-3), a loss, rather than a gain, of an rDNA repeat is expected. This model was proposed previously to explain the observed strong dependence of rDNA repeat expansion and contraction on FOB1 (18).