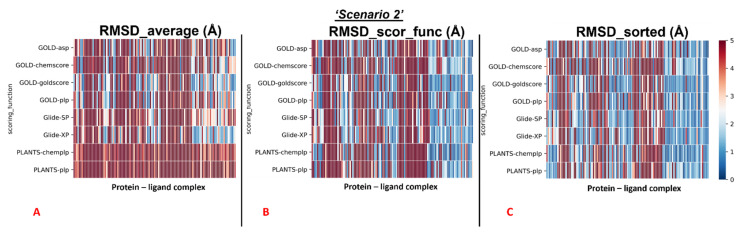
Figure 6.
Colormaps represent the results of the self-docking experiments in the case in which the crystallographic water molecules at 5 Å or nearer to the ligand itself are taken into account during the docking runs. (A) Results coming from the average of the RMSDs of all the poses for each docking run. (B) Results derived just from the RMSD between the crystallographic ligand coordinates and the pose classified as the best from the scoring function.(C)Results of the self-docking experiments if just the pose showing the best RMSD value between its coordinates and the crystallographic ones is retained. The x-axis lists all the different protein–ligand complexes, which are plotted against the different pairs docking program-scoring function used for this study, reported in the y-axis.