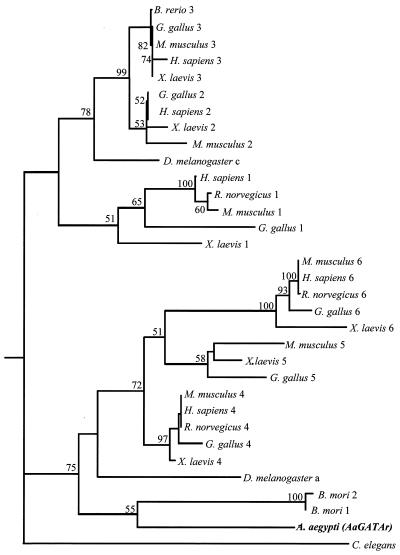
FIG. 3.
Dendrogram generated from amino acid sequences corresponding to DNA-binding domains of GATA transcription factors. The method of neighbor joining, using the distance formula of Kimura, was applied to generate the tree. Numbers at the nodes correspond to bootstrap values in 100 replicates. The accession numbers for the GATA sequences used are A. aegypti, AJ400338; B. mori 1, P52167; B. mori 2, P52167; Brachydanio rerio, Q91428; Caenorhabditis elegans 1, P28515; Drosophila melanogaster a, P52168; D. melanogaster c, D50542; Gallus gallus 1, P17678; G. gallus 2, P23824; G. gallus 3, P23825; G. gallus 4, P43691; G. gallus 5, P43692; G. gallus 6, P43693; Homo sapiens 1, P15976; H. sapiens 2, P23769; H. sapiens 3, P23771; H. sapiens 4, P43694; H. sapiens 6, Q92908; Mus musculus 1, P17679; M. musculus 2, AB000096; M. musculus 3, P23772; M. musculus 4, Q08369; M. musculus 5, P97489; M. musculus 6, S82462; Rattus norvegicus 1, P43429; R. norvegicus 4, P46152; R. norvegicus 6, P46153; Xenopus laevis 1, P23767; X. laevis 2, P23770; X. laevis 3, P23773; X. laevis 4, Q91677; X. laevis 5, P43695; X. laevis 6, Q91678.