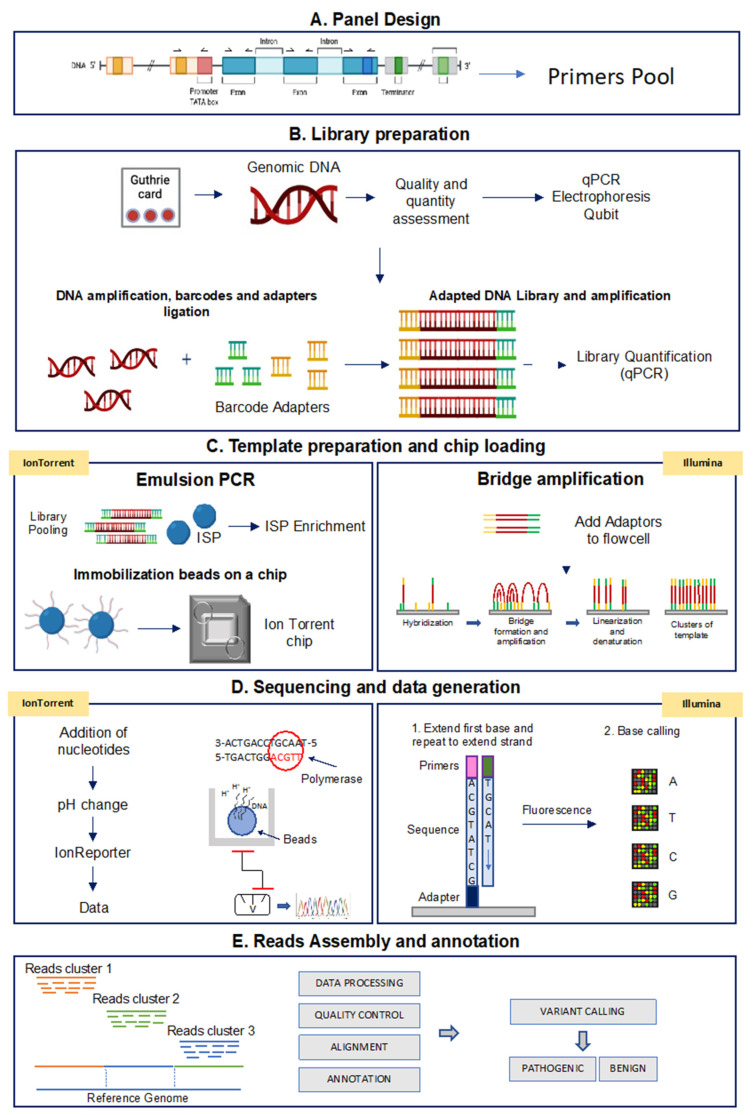
Figure 2.
Analytical workflow of targeted sequencing for the Ion Torrent and Illumina methods. (A) Panel design: the Designer software helps to create custom assays based on PCR target selection. (B) Library preparation: library construction is the preparation of the nucleic acid target into a form compatible with the sequencing system to be used. (C) Template preparation and chip loading: target enrichment is used in NGS workflows to capture only genomic DNA regions of interest. (D) Sequencing and data generation: IonTorrent platform: microwells of the chip is flooded by nucleotides that when binding to the complementary nucleotide on a template, release an ion. At each flow, the electrical signal at each well is measured, indicating that a reaction has occurred; Illumina platform: the fragments are clonally amplified on the slide utilizing fluorescently labeled reversible-terminator nucleotides; (E) Read assembly and annotation: starting from Binary Alignment Map (BAM) and Variant Call Format (VCF) files, variants are prioritized based on localization, functional effect, mode of inheritance, coverage and Minor Allelic Frequency (MAF) to obtain disease-correlated variants.