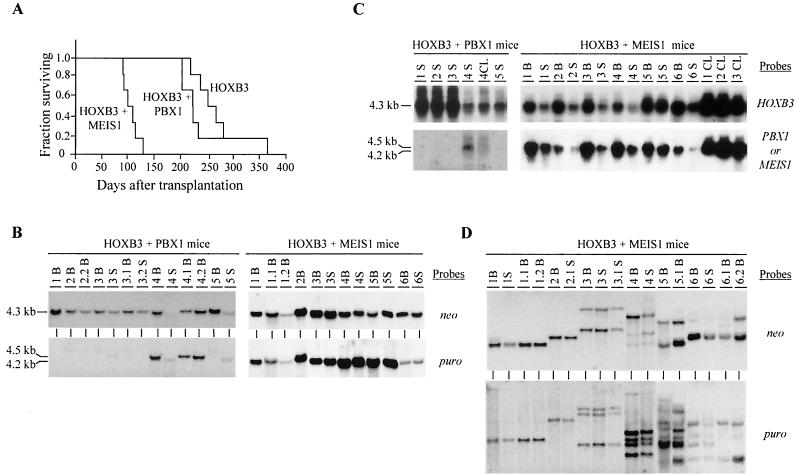
FIG. 2.
Demonstration of collaboration between HOXB3 and MEIS1, but not HOXB3 and PBX1, in leukemogenesis. (A) Survival graph demonstrating the collaboration between HOXB3 and MEIS1, but not PBX1, in the development of AML. The survival of the HOXB3-MEIS1 mice was significantly shorter than that of the HOXB3 mice (P < 0.001, two-tailed Student's t test) and the HOXB3-PBX1 mice (P < 0.007). The survival of the HOXB3-PBX1 mice was not significantly different from that of the HOXB3 mice. (B) Southern blot analyses of genomic DNA isolated from the bone marrow and/or spleen of the HOXB3-PBX1 and HOXB3-MEIS1 chimeras. DNA was digested with KpnI to release the integrated HOXB3 (4.3-kb), MEIS1 (4.2-kb), or PBX1 (4.5-kb) proviral fragments. The membranes were hybridized with a neo-specific probe to detect the HOXB3 provirus and a puro-specific probe to detect the MEIS1 or PBX1 provirus. (C) Northern blot analysis of total RNA (10 μg) isolated from bone marrow or spleen cells of the HOXB3-PBX1 and HOXB3-MEIS1 mice. The membranes were hybridized with full-length HOXB3, MEIS1, or PBX1 cDNA probes. (D) Southern blot analysis of DNA isolated from bone marrow of primary and secondary HOXB3-MEIS1 mice. The DNA was digested with EcoRI, which cuts the integrated provirus once, thus generating a unique fragment for each proviral integration site. The membranes were hybridized first with a neo-specific probe for detection of the HOXB3 proviral fragment(s) (top panel) and then subsequently with a puro-specific probe to detect the MEIS1 proviral fragment(s) (bottom panel). In panels B, C, and D, each primary recipient is identified with a specific number and its secondary recipients or cell lines generated from each primary recipient, with a derivative thereof (e.g., 1.1 and 1.2 and CL1, CL2, etc.). B, bone marrow; S, spleen; CL, cell lines.