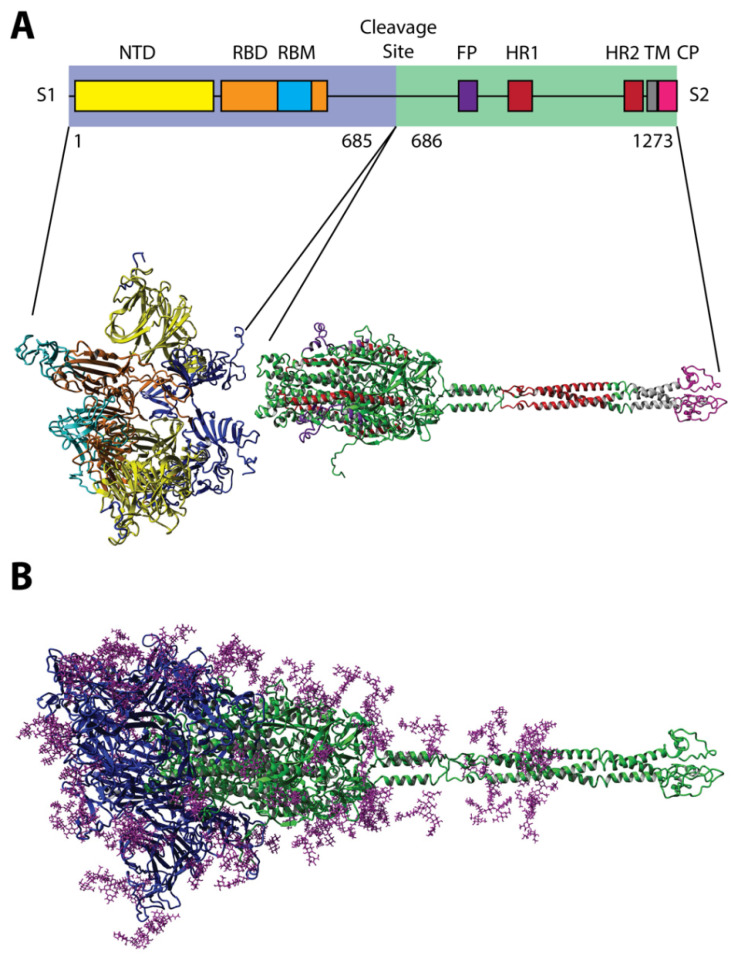
Figure 1.
Three-dimensional predicted model of the spike (S) glycoprotein of the SARS-CoV-2 virus composed of (A) two well-defined structural domains (S1 and S2), decorated with (B) 22 N-glycan residues as modeled using 6VSB and 6VXX [38,39]. Monomers of the S protein, composed of polypeptide chains of 1273 amino acids, form homotrimer spikes on the virus surface [18]. Spike protein monomers are composed of three major structural domains: head, stalk, and cytoplasmic tail. The head comprises the N-terminal domain (NTD; yellow) and the receptor-binding domain (RBD; orange), which displays the receptor-binding motif (RBM; cyan) that is responsible for interaction with cell receptors [40]. RBDs in non-activated viral S glycoprotein trimers are present in a hidden “down” conformation. The S glycoprotein is cleaved by host proteases (trypsin and furin) at the site between the S1 and the S2 subunits [41,42]. The S2 domain of the S protein consists of fusion peptide (FP; purple), two heptad-repeat domains (HR1 and HR2; red), a transmembrane domain (TM; gray), and a cytoplasm domain (CP; pink). A second proteolytic site (S2′ site), located within the S2 subdomain, is also cut by type II transmembrane serine protease (TMPRSS2) as well as cathepsin B and L (CatB/L) to enable virus-cell fusion by triggering the dissociation of S1 and the irreversible refolding of S2, a conformational change of the S protein and the fusion of the viral envelope and endosomes [42].