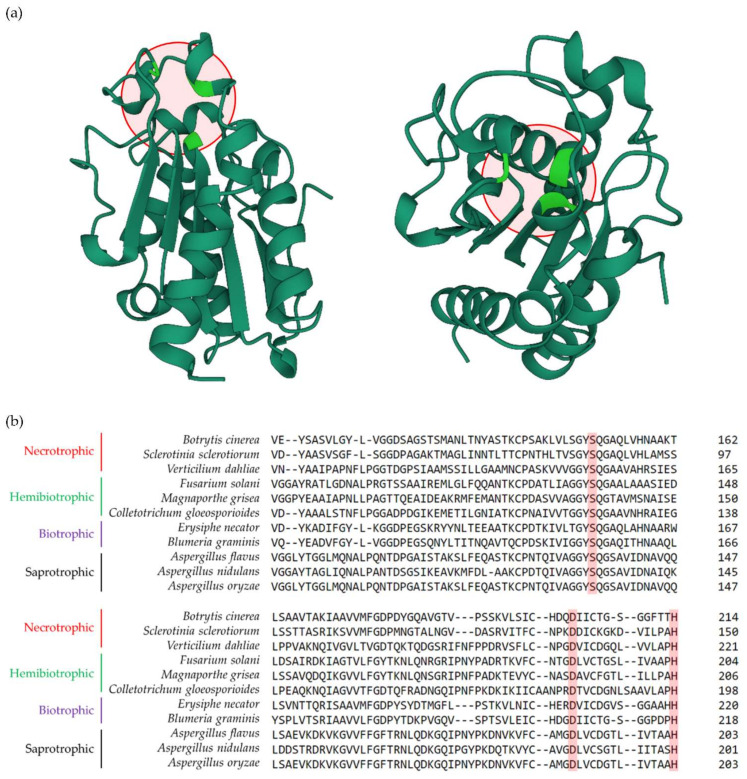
Figure 1.
Structure of the Fusarium solani cutinase and the conserved catalytic triad among fungal species with different lifestyles. (a) The crystal structure of the Fusarium solani CUT enzyme (protein ID: 1CEX) according to the RCSB Protein Data Bank (PDB) (https://www.rcsb.org/ accessed on 20 December 2021). Original structure was elucidated based on X-ray crystallography analyses at atomic resolution of 1.0A° performed by [22]. The structure can be seen in a front (left) and upright (right) overviews. Red circles show the catalytic triad consists of Ser, Asp and His, that are marked in green. (b) Protein alignment of cutinases isolated from fungal species with different lifestyles, including necrotrophic (e.g., Botrytis cinerea, Sclerotinia sclerotiorum and Verticilium dahliae), hemibiotrophic (e.g., Fusarium solani, Magnaporthe grisea and Colletotrichum gloeosporioides), biotrophic (e.g., Erysiphe necator and Blumeria graminis), and saprotrophic (e.g., Apergillus flavus, Aspergillus nidulans and Aspergillus oryzae). The conserved amino acids of Ser, Asp and His that build the catalytic triad are highlighted in red rectangles. Protein sequence alignment was performed using the tools embedded in the ClustalW2 software (https://www.ebi.ac.uk/Tools/msa/clustalw2/ accessed on 12 February 2022).