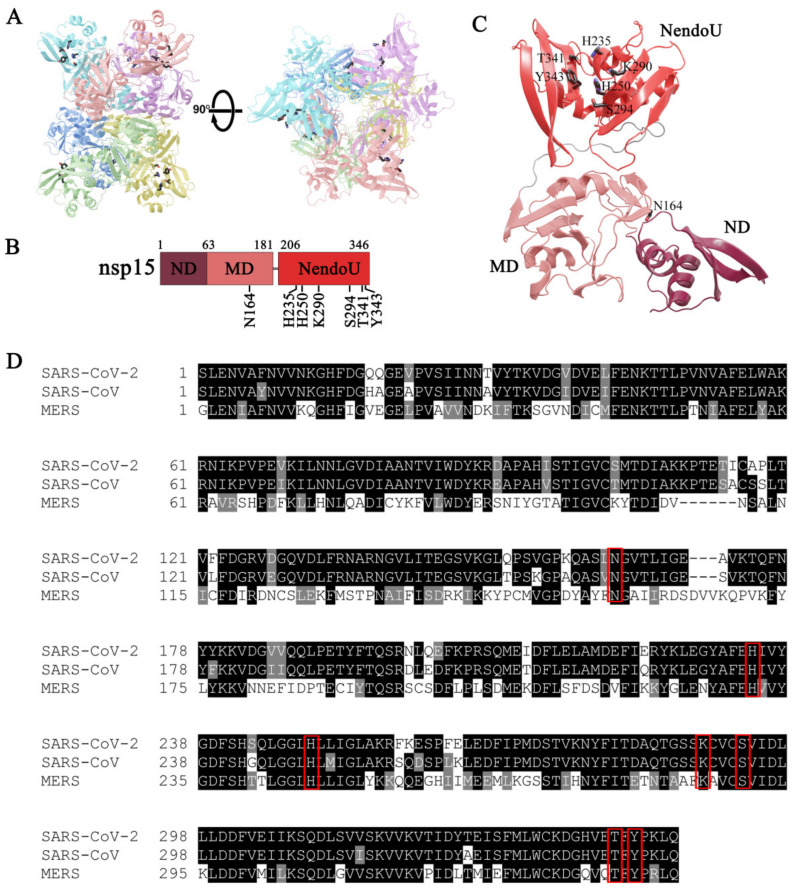
Figure 1.
(A) Orthogonal views of the hexameric form of nsp15, which is formed by dimers of trimers. Monomers are colored in cyan, pink, violet, blue, green and yellow. Active site residues are shown as sticks. (B) nsp15 domain organization. ND, amino acids (aa) 1–63; MD, aa 64–181; NendoU, aa 206–346. The location of the conserved residues is represented. (C) Localization of the nsp15 domains in the monomer (PDB code 6W01). Mutated residues are shown as sticks. (D) Sequence alignment of nsp15 from SARS-CoV-2 (UniProt ID: P0DTD1), SARS-CoV (UniProt ID: P0C6X7) and MERS-CoV (YP_009047225). Residues are colored according to their conservation: in black, residues that are identical; in grey, residues that are not identical but have similar properties; in white, residues that are different. Residues mutated in this work are highlighted with a red box.