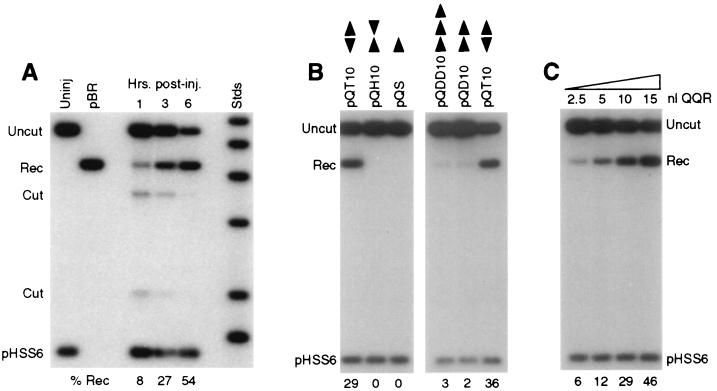
FIG. 2.
Cleavage and recombination in oocytes. (A) Time course of cleavage and recombination in Xenopus oocytes after injection of QQR. Circular pQT10 DNA (0.3 fmol; this corresponds to 0.6 fmol of binding sites) was injected into oocyte nuclei, following the scheme shown in Fig. 1B; 10 nl (30 fmol) of a solution of QQR was delivered to each oocyte, and DNA was recovered at the indicated times after enzyme injection. Recovered DNA was analyzed as described in the legend to Fig. 1B; the positions of the expected fragments (Uncut, Cut, and Rec) are indicated. pHSS6 is a circular plasmid that was included as a recovery control (22). Uninjected pQT10 (Uninj) is shown in the first lane, and uninjected pBR322 (pBR) serves as a marker for the position of recombination products. Stds, linear size standards. The percentage of the recovered DNA that was in the product band (Rec) is indicated below each lane. (B) Effect of recognition site disposition on cleavage and recombination. The number and orientation of sites are indicated by the arrowheads above each lane, with the point designating the A end of the binding site. pQS has a single recognition site; pQT10 has two sites in tail-to-tail inverted orientation; the two sites in pQH10 are in head-to-head orientation; pQD10 has two sites in direct repeat orientation; and pQDD10 has three direct repeats. Each pair of neighboring sites is separated by 10 bp. DNA and enzyme concentrations were as in panel A; incubation was done overnight. (C) Cleavage and recombination in oocytes after injection of various amounts of QQR (3 fmol/nl), as indicated. The DNA substrate was pQT10, and incubation was overnight.