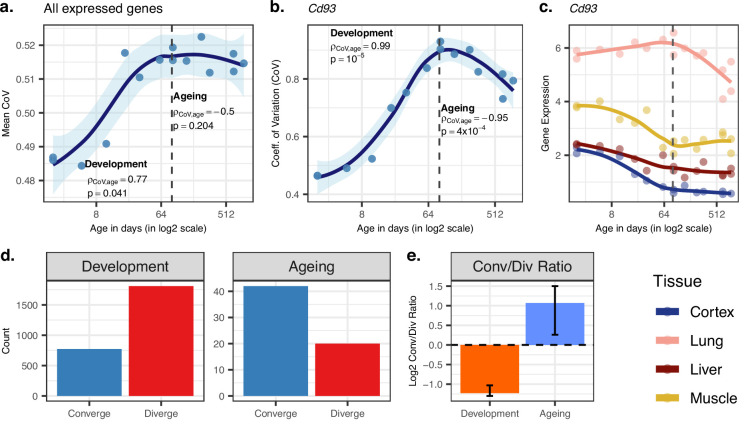
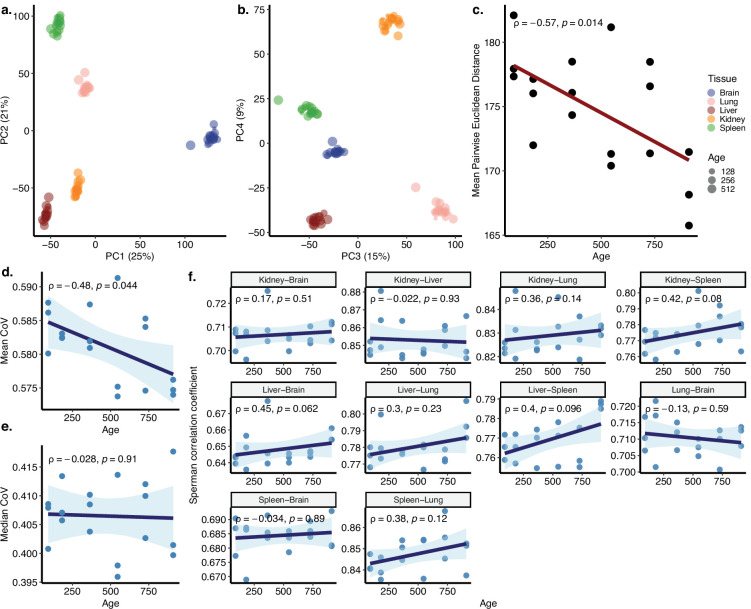
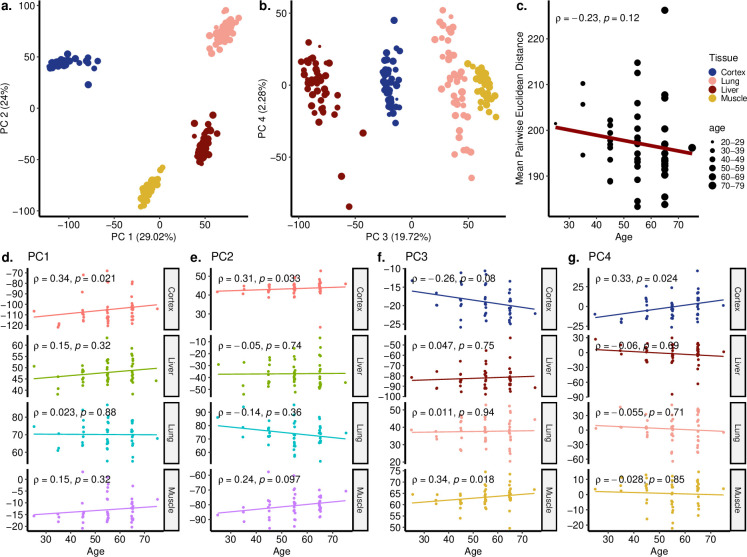
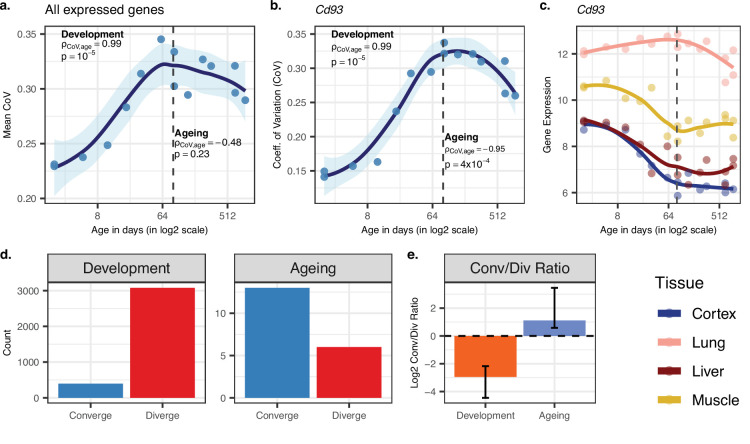
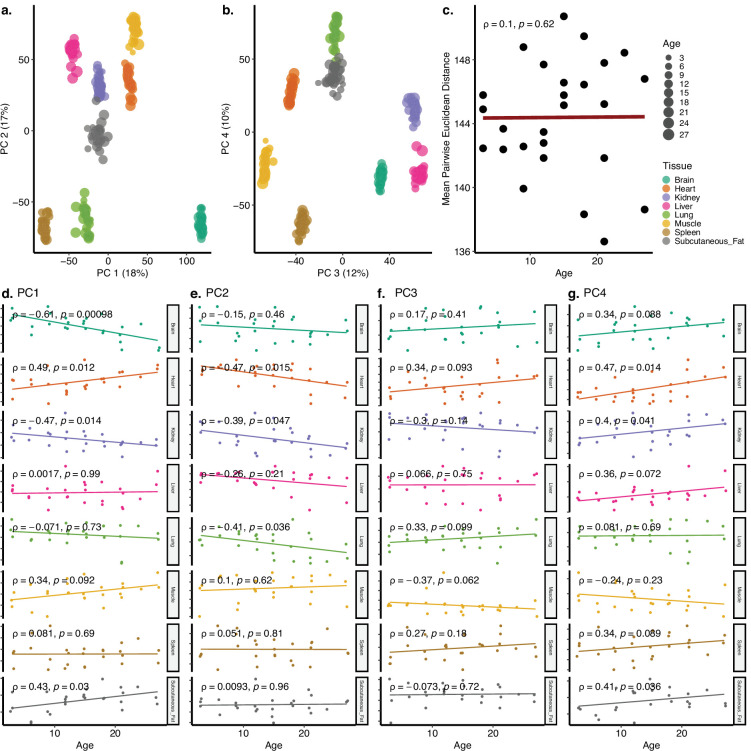
Figure 2. Age-related change in gene expression variation among tissues estimated with coefficient of variation (CoV).
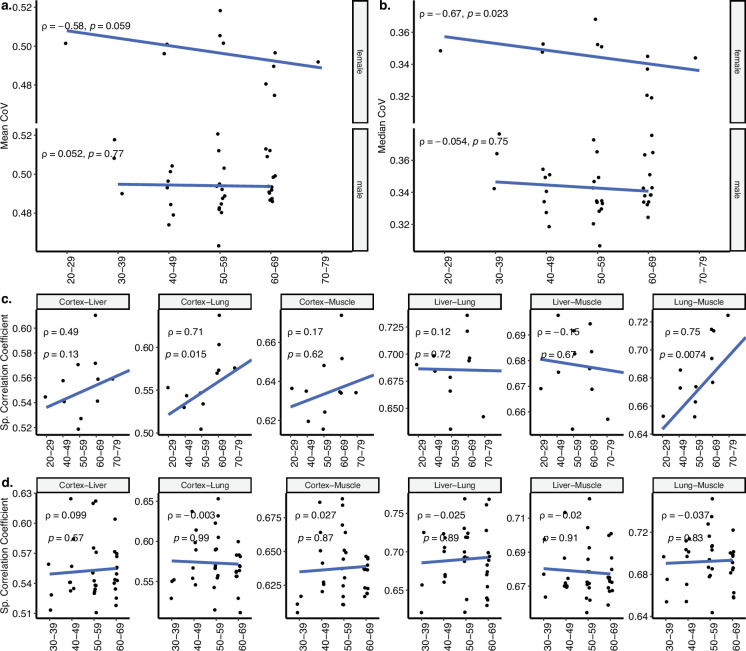
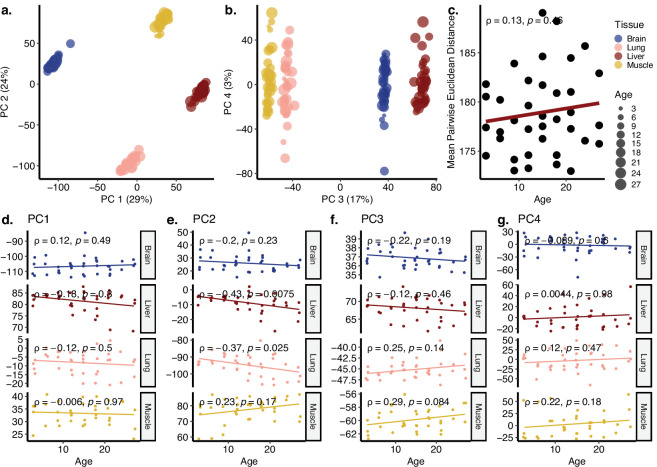
(a) Transcriptome-wide mean CoV trajectory with age. Each point represents the mean CoV value of all protein-coding genes (15,063) for each mouse (n = 15) except the one that lacks expression data in the cortex. (b) Age effect on CoV value of the Cd93 gene which has the highest rank for the divergence-convergence (DiCo) pattern in four tissues (Materials and methods). CoV increases during development and decreases during ageing, indicating expression levels show DiCo patterns among tissues. (c) Expression trajectories of the gene Cd93 in four tissues. (d) The number of significant CoV changes with age (false discovery rate [FDR]-corrected p-value<0.1) during development (left, nconverge = 772, ndiverge = 1809) and ageing (right, nconverge = 42, ndiverge = 20). Converge: genes showing a negative correlation (ρ) between CoV and age; diverge: genes showing a positive correlation between CoV and age. (e) Log2 ratio of convergent/divergent genes in development and in ageing. The graph represents only genes showing significant CoV changes (FDR-corrected p-value<0.1, given in panel d). Error bars represent the range of log2 ratios calculated from leave-one-out samples using the jackknife procedure (Materials and methods, values are given in Figure 2—source data 1).
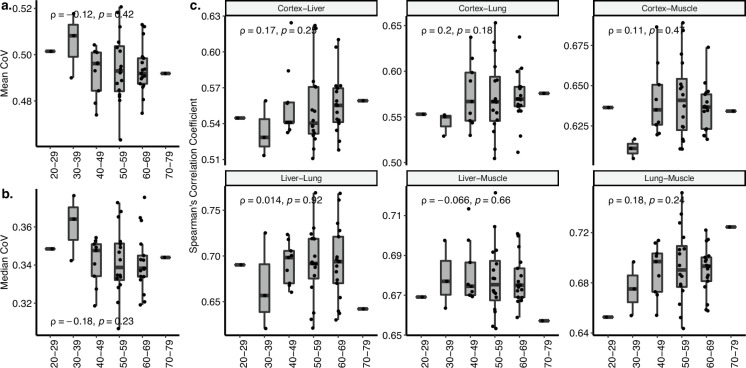
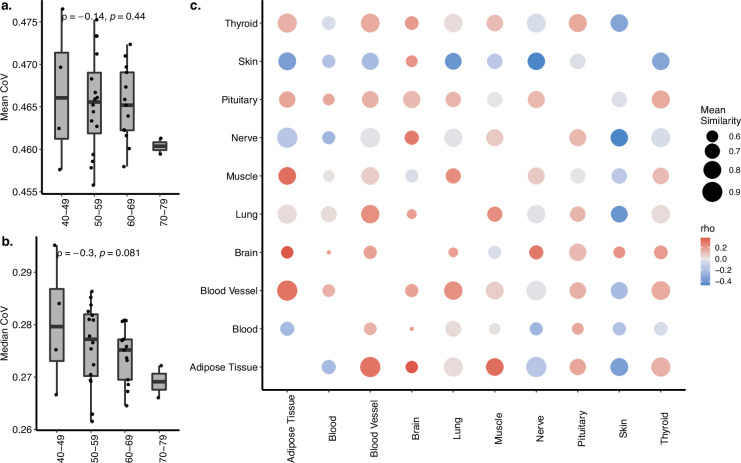
Figure 2—figure supplement 1. Age-related change in coefficient of variation (CoV) summarised across genes using median CoV values.
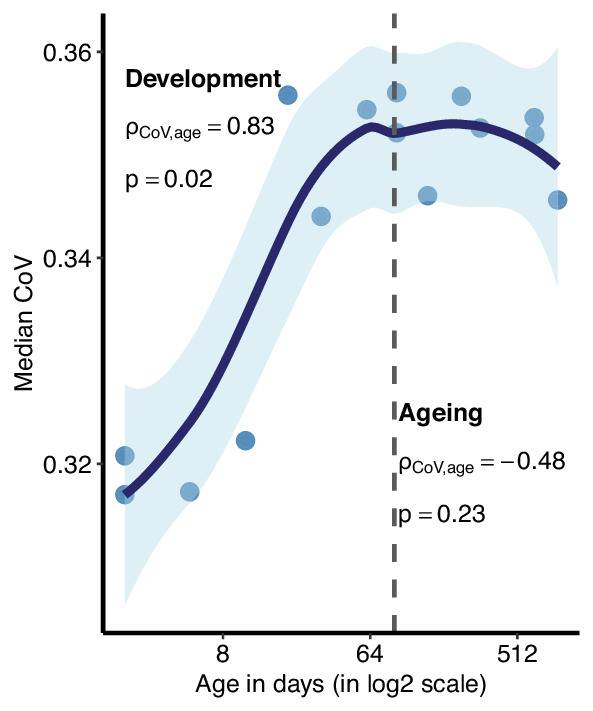
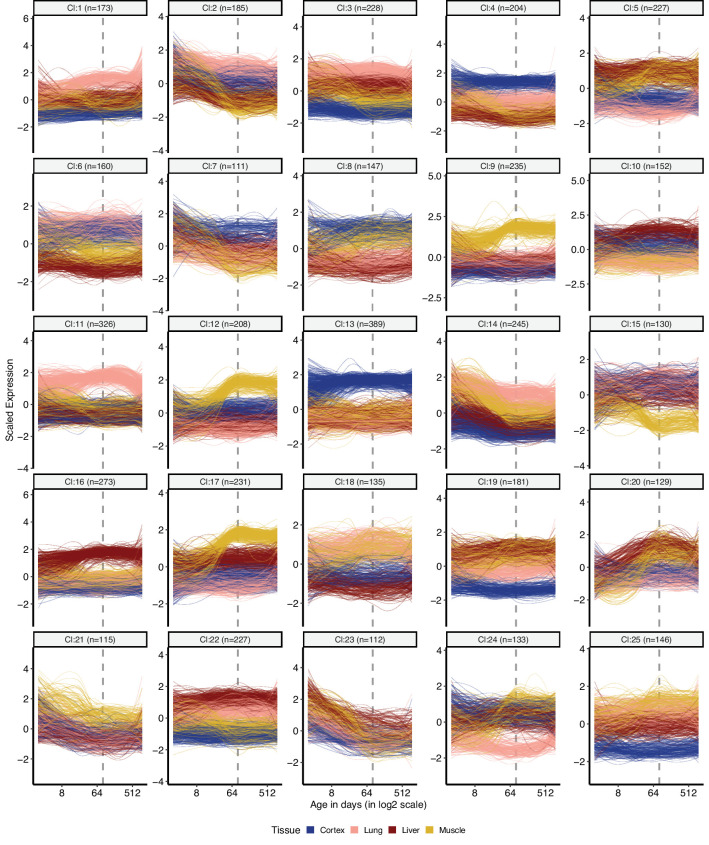
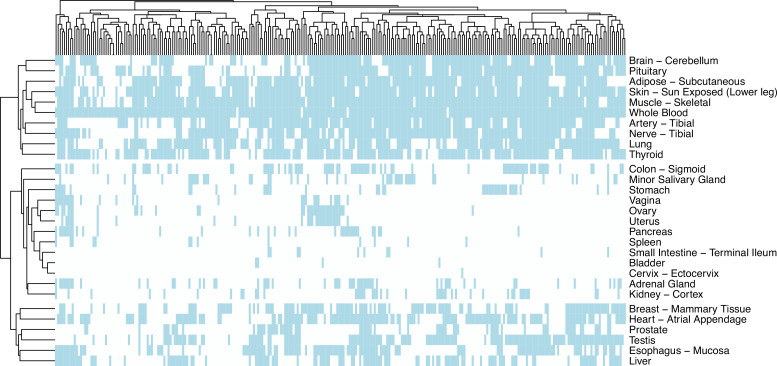
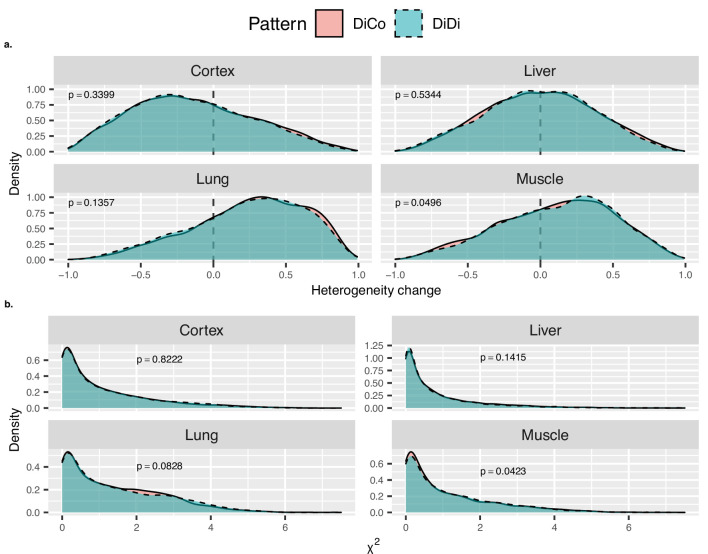
Figure 2—figure supplement 2. Clustering of divergence-convergence (DiCo) genes by expression variations (coefficient of variation [CoV]) among tissues.
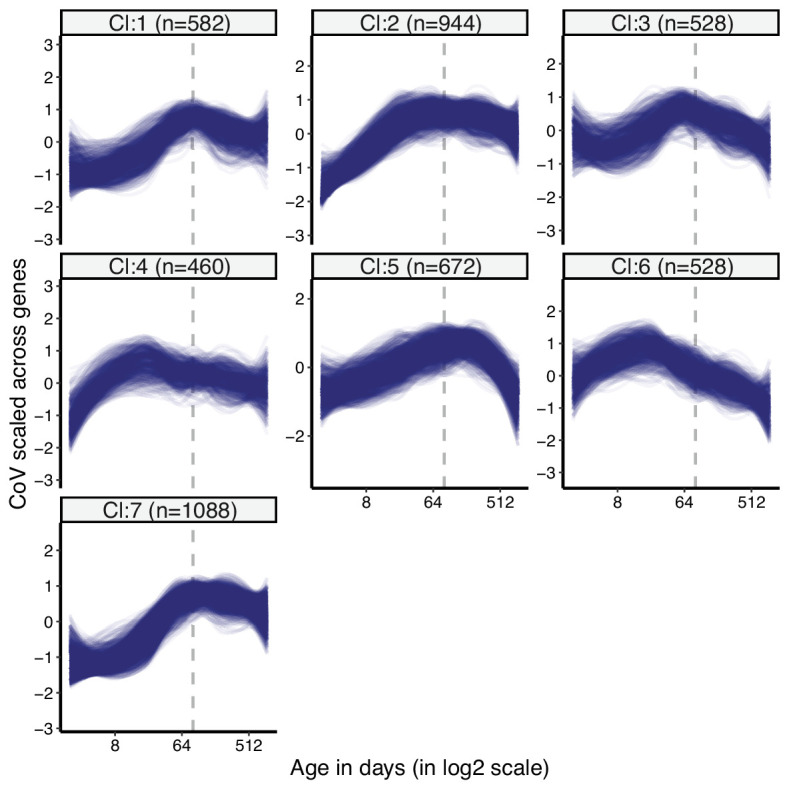
Figure 2—figure supplement 3. Clustering of divergence-convergence (DiCo) genes by expression levels in tissues.
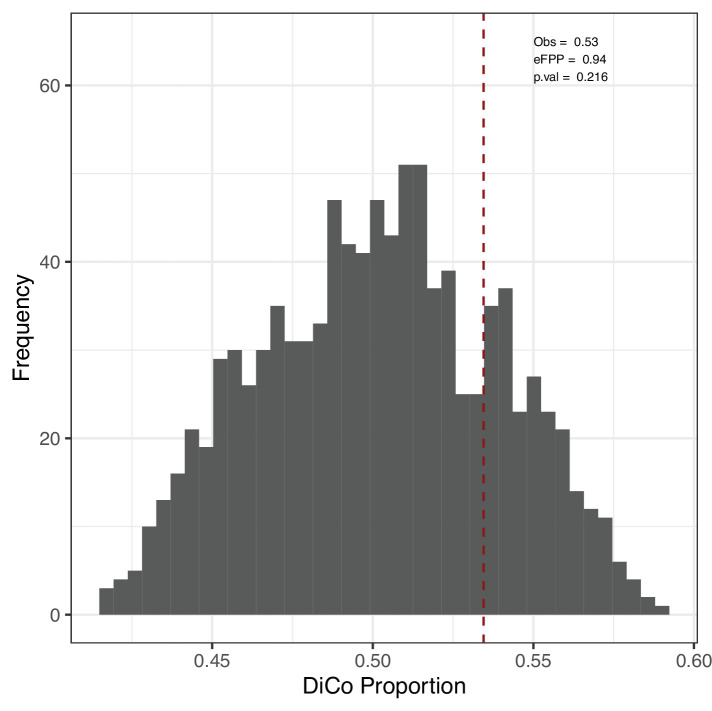
Figure 2—figure supplement 4. Number of genes with inter-tissue divergence and convergence tendencies in development and ageing.
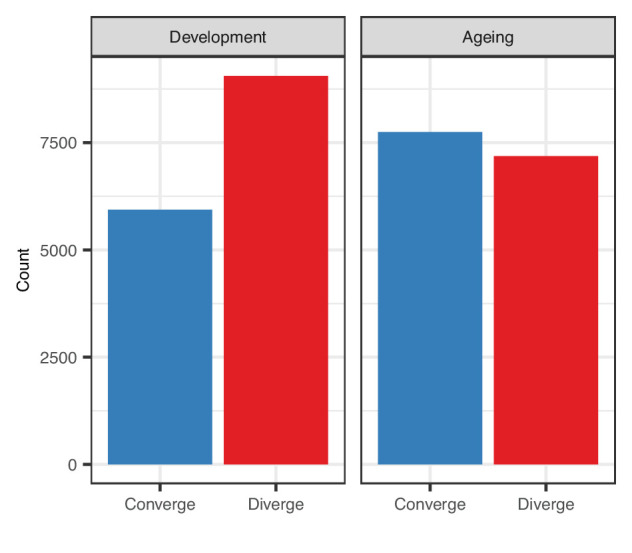
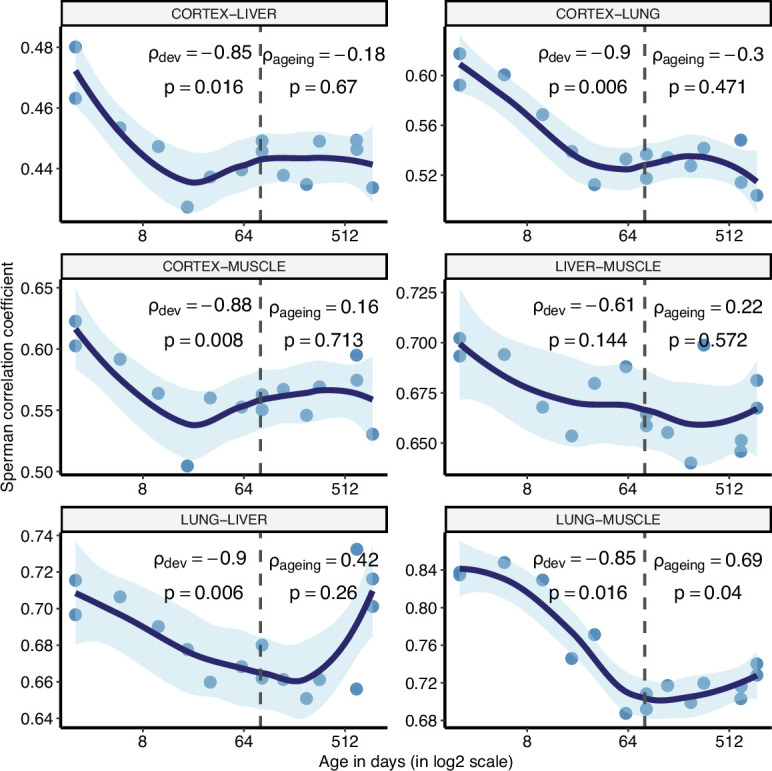
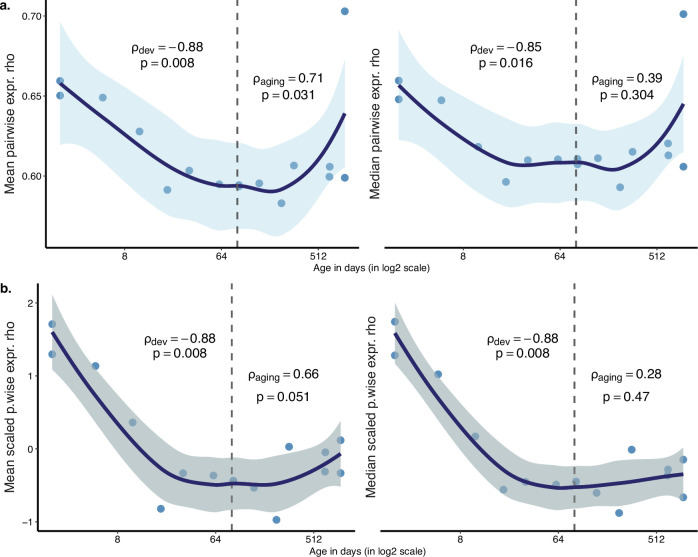
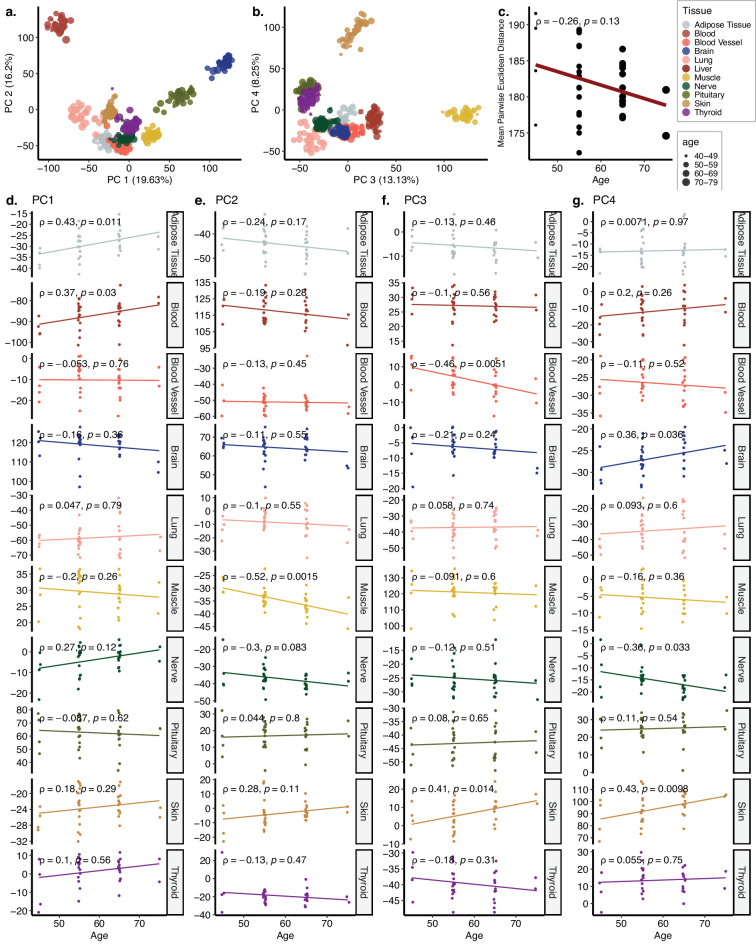
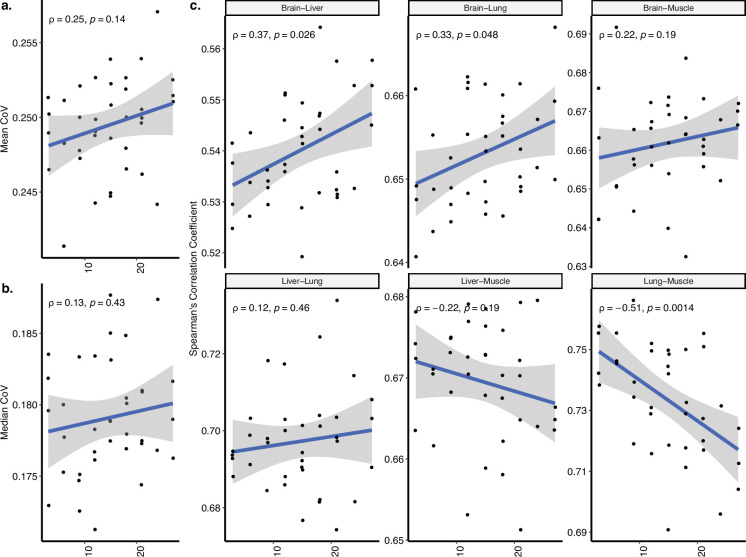
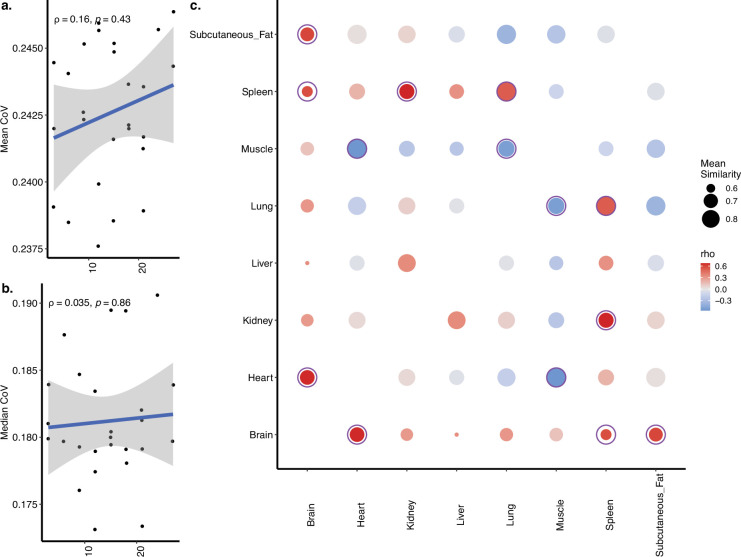
Figure 2—figure supplement 5. Pairwise tissue expression correlations.