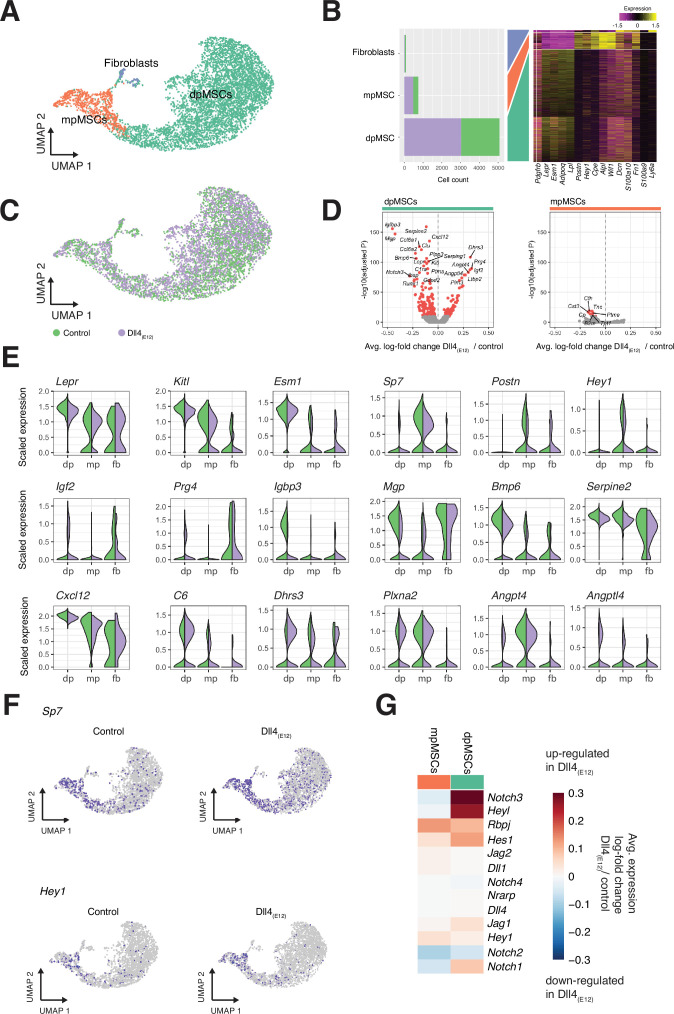
Figure 9. Single cell RNA sequencing (scRNA-seq) analysis of mesenchymal stromal cell (MSC) subclusters.
(A) UMAP projection of MSCs in both control and pLIVE-Dll4(E12) samples colored by Louvain clusters. Metaphyseal MSCs (mpMSCs), diaphyseal MSCs (dpMSCs), and fibroblasts are indicated. (B) Barplot showing absolute numbers of cells per sample (left), and scaled expression heatmap of the top 10 marker genes (right) in each of the clusters shown in (A). (C) UMAP projection of all cells colored by experimental condition green = control, purple = Dll4(E12). (D) Differential expression volcano plots showing -log10(adjusted p-value) against average log-fold change of treatment/control for dpMSCs (left) and mpMSCs (right). Genes with an adjusted p-value less than 1e-10 are colored in red. (E) Violin plots showing selected differentially expressed genes in the three MSC subpopulations. Each violin is split along the vertical axis into control and pLIVE-Dll4(E12). (F) UMAP projections of control and Dll4(E12)-treated cells colored for the expression of Sp7 and Hey1. (G) Heatmap of the average expression log-fold change of selected Notch pathway genes with a p-value < 1e-10 and log-fold change >0.2 in dpMSCs and mpMSCs.