Fig 3. Gene modules co-regulated by infliximab and associated with subsequent improvements in psychomotor speed in treatment resistant depression (TRD) are enriched for pathways related to oxidative stress, mitochondrial degradation, inflammation, and cancer metabolism.
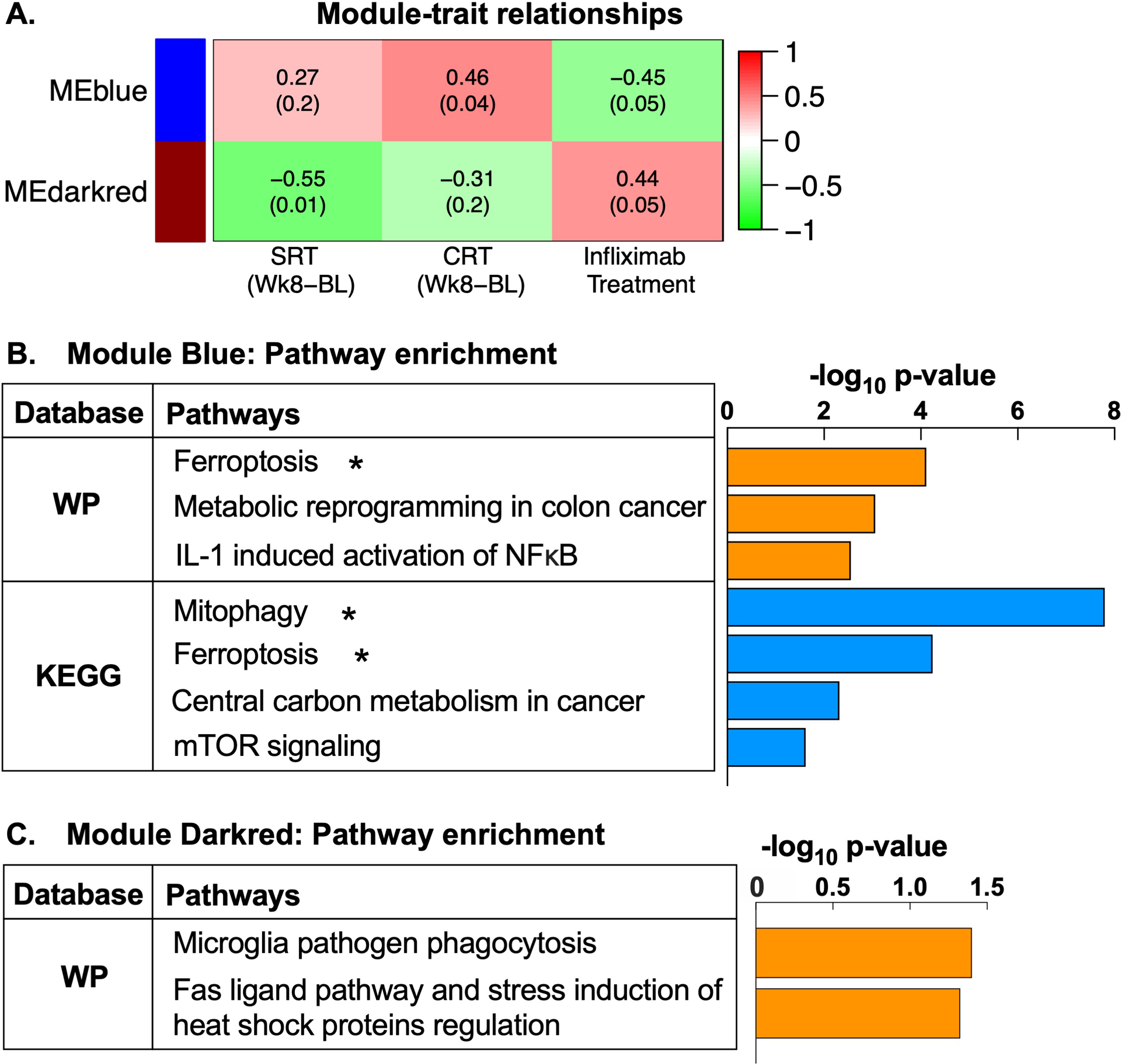
Among TRD patients with high inflammation (n=20 patients), change in gene expression following a single infusion of infliximab or placebo (Week 2 post-infusion - baseline) clustered into 33 differential gene co-expression modules, labeled by distinct color names, and constituent genes summarized by a module eigengene (ME). Of these, MEs of two gene modules, blue and darkred, were associated with both infliximab treatment and subsequent change in simple or choice reaction time (SRT and CRT) at Week 8 (A). Treatment with infliximab was associated with decreasing MEblue (r=−0.45, p=0.049), and lower values of MEblue corresponded to more negative values of change in CRT (indicating faster reaction speed at Week 8 relative to baseline) (r=0.46, p=0.04). Conversely, infliximab positively correlated with MEdarkred (r=0.44, p=0.05), and larger values of MEdarkred in turn correlated with more negative values of change in SRT (hence greater improvement) (r=−0.55, p=0.01). Functional annotation of the gene transcripts in Module Blue (n=1440) within WikiPathways and KEGG databases revealed significant enrichment of pathways related to oxidative stress and mitochondrial degradation (ferroptosis and mitophagy), as well as immune function (interleukin-1-induced activation of NF-kappa-B) and cancer metabolism (metabolic reprogramming in colon cancer, central carbon metabolism in cancer) (all p<0.05) (B). Gene transcripts within Module darkred (n=150) enriched immune pathways including microglia pathogen phagocytosis and a Fas ligand pathway (all p<0.05) (C). ME, module eigengene; SRT, simple reaction time; CRT, choice reaction time; WP, WikiPathways; KEGG, Kyoto Encyclopedia Gene and Genome; IL-1, interleukin-1; NFκB, nuclear factor kappa-B; mTOR, mammalian target of rapamycin. *pathways are significantly enriched at p<0.05 and q<0.1.
