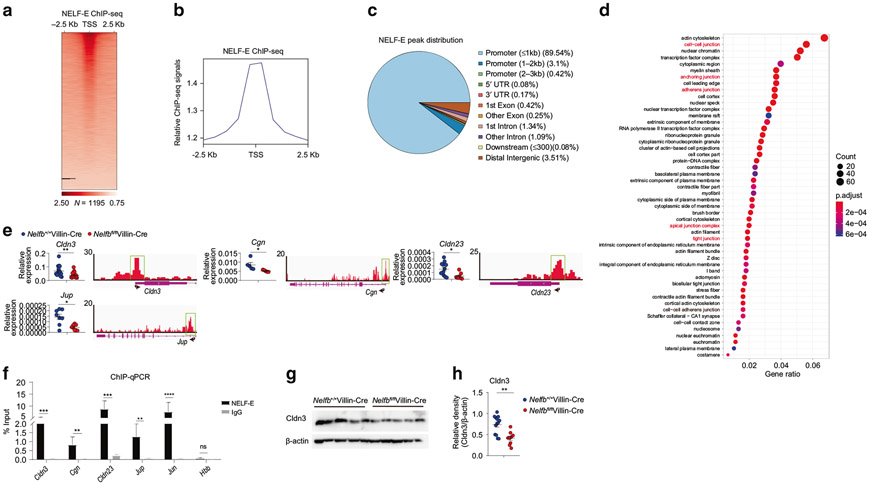
Fig. 5. Epithelial NELF promotes the expression of a subset of cell junction-related genes with direct NELF binding to their promoter regions.
a Heat map of NELF-E ChIP-seq signals around TSS regions for colonic IEC-expressed genes in Nelfb+/+Villin-Cre mice at steady state. Each row indicates one gene sorted by the peak heights in Nelfb+/+Villin-Cre mice. The signals were normalized to input signals. The number of peaks in the whole genome is shown at the bottom. b Relative NELF-E ChIP-seq signals around TSS regions as (a) shown. c The genomic distribution of NELF-E ChIP-seq peaks. d GO analysis of NELF-E ChIP-seq peaks for cellular component classes and many cellular components (red font) are related to epithelia cell–cell junction. e qPCR analysis of epithelia cell junction-related genes expression in Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice (n = 6–12). Tracks of NELF-E ChIP-seq are shown for these genes. f The occupancies of NELF-E in the TSS region of cell junction-related genes were assessed by ChIP-qPCR in Nelfbfl/flVillin-Cre versus Nelfb+/+Villin-Cre mice. g, h Immunoblotting analysis (g) and quantification (h) of Cldn3 in colon tissues of Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice (n = 9–12). Data are pooled from at least two (e, f, and h) independent experiments and shown as mean ± SEM. Student’s t-test was performed; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; ns not significant (p > 0.05). Images (g) represent at least two independent experiments.