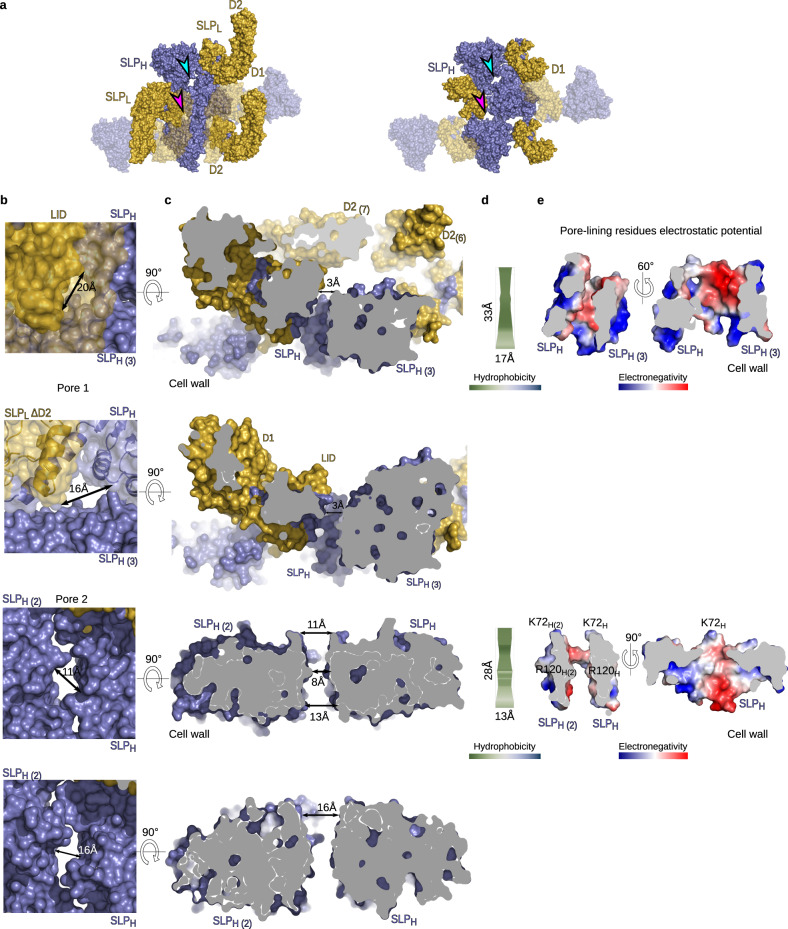
Fig. 5. C. difficile S-layer is a tightly packed array with very narrow pores.
a Surface representation of wild-type H/L (7ACY, left) and SlpARΔD2 H/L (7ACZ, right) crystal packing showing pores in the 3D crystal lattice. Positions of pores marked with arrowheads (pore 1 in magenta, pore 2 in cyan) are equivalent in both lattices. b Zoomed-in view of the pores generated by H/L multimerization. Pore 1, top view covered by D2 in SlpACD630 (first panel) and in SlpARΔD2 (second panel). Pore 2 – top view for SlpACD630 (third panel) and SlpARΔD2 (fourth panel). Widest openings are labelled for each pore. Arrows indicate the widest points in each pore, which are exposed in SlpARΔD2 due to the lack of D2 that completely covers it in SlpACD630. c Cross-section views of pore 1 and pore 2 in SlpACD630 (first and third panels, respectively) and SlpARΔD2 (second and fourth panels, respectively). Neighbouring SLPH (slate blue) and SLPL (gold) molecules that create the pores are shown in surface representation. d Hydrophobicity characteristics of the residues lining pore 1 (top) and 2 (bottom) calculated in ChexVis (see Methods section for details) according to Kyte-Doolittle scale, ranging from hydrophilic (green) to hydrophobic (blue), as per hydrophobicity gradient key. e Poisson–Boltzmann electrostatic potential calculated for residues lining pore 1 (first panel) and 2 (second panel) in SlpACD630 represented as a charge distribution (positive in blue and negative in red, as per electronegativity gradient key). Views and scale are as in c (left) and as a slice across the largest pore surface (right). Pseudo-symmetry-related lysine residues at the top and arginine residues at the bottleneck of pore 2 are highlighted.