Abstract
Multimerin-1 (MMRN1) is a platelet protein with a role in haemostasis and coagulation. It is also present in endothelial cells (ECs) and the extracellular matrix (ECM), where it may be involved in cell adhesion, but its molecular functions and protein–protein interactions in these cellular locations have not been studied in detail yet. In recent years, MMRN1 has been identified as a differentially expressed gene (DEG) in various cancers and it has been proposed as a possible cancer biomarker. Some evidence suggest that MMRN1 expression is regulated by methylation, protein interactions, and non-coding RNAs (ncRNAs) in different cancers. This raises the questions if a functional role of MMRN1 is being targeted during cancer development, and if MMRN1’s differential expression pattern correlates with cancer progression. As a result, it is timely to review the current state of what is known about MMRN1 to help inform future research into MMRN1’s molecular mechanisms in cancer.
Keywords: biomarkers, cancer, molecular basis of health and disease, transcription
Background
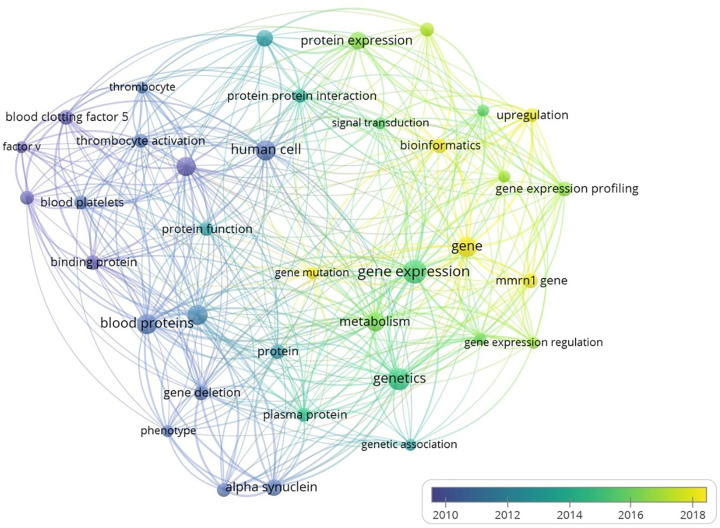
Multimerin-1 (MMRN1) is a member of the EMILIN/multimerin family of proteins, found in platelets (α-granules of resting platelets), megakaryocytes, endothelial cells (ECs, Weibel–Palade bodies) and the extracellular matrix (ECM) [1–6]. In response to specific triggers, MMRN1 is secreted from platelet α-granules and Weibel–Palade bodies of ECs [7–9]. MMRN1 platelet-related functions include platelet adhesion [5,10–13], factor V regulation [14–18], and MMRN1 deficiency is associated with bleeding risks in Quebec platelet disorder [19–21]. These functions, however, shall not be covered in the current article as these have already been reviewed [22]. Instead, the focus will be on MMRN1’s non-platelet-related functions, including mmrn1 differential gene expression and its proposed use as a cancer biomarker [23–25]. A bibliometric network analysis illustrates how the research on MMRN1’s platelet-related functions i.e., ‘blood’, ‘platelets’, ‘factor V’, has shifted to the analysis of MMRN1 ‘gene expression’, ‘protein expression’, using ‘bioinformatics’ over recent years (Figure 1). The reports on MMRN1’s differential gene expression and also highlight the lack of molecular studies aimed to describe MMRN1’s physiological functions.
Figure 1. Bibliometric network analysis of MMRN1 in the scientific literatures.
The network analysis was carried out using VosViewer (https://www.vosviewer.com, [202]). Over time, the research focus has shifted from MMRN1’s role in platelets (purple, blue) to its differential gene expression (green, yellow) in recent years.
Whereas the role of other EMILIN/multimerin family members in cancer has been the focus of various studies [26–37], a molecular mechanism for MMRN1 is yet to be described. It is also unclear if MMRN1’s functions are indiscriminatory of its cellular locations, platelets, ECs and the ECM, which all play roles in cancer. Platelets are associated with metastasis [38–40]. The interaction with platelets, facilitates circulating cancer cells to evade natural killer cells and adhere to vascular walls. The cancer cells can subsequently cross the vascular endothelium and exit the circulation (extravasation), which can lead to metastasis [39–41]. The recruitment of the ECM and granulocytes by activated platelets drives the formation of the early metastatic niche that allows the cancer to survive and proliferate [39]. Activated platelets also release pro-angiogenic growth factors (e.g., vascular endothelial growth factor (VEGF)) which regulate tumour angiogenesis [41]. EC migration and proliferation are activated by VEGF [42] and interaction between ECs and cancer cells can further promote angiogenesis [43]. The ECM is involved in various processes including epithelial-to-mesenchymal transition and metastasis [44,45], the latter being an example of cancer plasticity [46]. The tumour ECM, which is mainly derived from tumour-associated fibroblasts, is denser and stiffer and has an altered, tumour-specific molecular expression profile, which alters cell–ECM interactions and signalling cascades to support tumour growth. Cancer plasticity is associated with changes in gene expression patterns, which is why MMRN1’s differential expression pattern in cancer is of great interest.
MMRN1 expression is significantly downregulated in 17 (bladder, breast, colon, oesophagous, liver, lung (adenocarcinoma and squamous cell carcinoma), ovary, prostate, rectum, renal (cell carcinoma and papillary cell carcinoma), skin, stomach, testis, thyroid, uterus (uterine carcinosarcoma and uterine corpus endometrial carcinoma)) out of 22 cancer types, and significantly upregulated in acute myeloid leukaemia and pancreatic cancer (TNM plot pan-cancer analysis [47]). MMRN1 is also expressed by various cancer cell lines. Despite such analyses and clinical observations of MMRN1 differential expression in cancer [24,25], the molecular mechanisms driving these changes are currently unknown and raises various questions: is MMRN1 differential expression merely a result of other cellular changes or is it regulated by tumour cells to aid the disease development and/or progression? How is MMRN1 expression regulated in cancer? What are MMRN1 functions in different cell types? Can MMRN1 expression levels help diagnose cancer and/or cancer stages? Does MMRN1 interact with tumour cells via protein–protein interactions? This review is intended to provide an overview of the state-of-the-art in this field, with the aim to provide information towards addressing these questions in future.
MMRN1 protein–protein interactions and their possible role in cancer
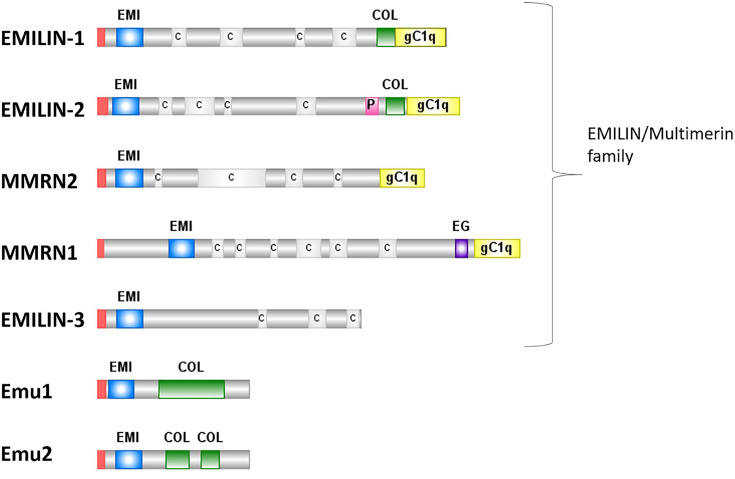
MMRN1 is a large glycoprotein which contains an N-terminal EMI-domain, an epidermal growth factor (EGF)-like domain, coiled-coil, and a C-terminal gC1q domain [6,48] (Figure 2). Some domains are shared between different family members (EMI, gC1q) but their functions vary (Figure 3). The EMI domain of EMILIN-1 and EMILIN-3 has been shown to regulate TGF-β signalling [49,50], whereas in EMILIN-2 the EMI domain regulates Wnt signalling in breast cancer [31]. The EMI domain also mediates protein–protein interactions including the interaction between the EMILIN-1 EMI domain and the EMILN-2 gC1q domain [48,51], and heparin binding to EMILIN-3 [50]. The EMI domain of MMRN1 is unique amongst family members as it has six cysteine residues instead of seven [52]. If the MMRN1 EMI domain is also involved in protein–protein interactions and signalling events as its family members is unclear.
Figure 2. The protein domains of the EMILIN/multimerin family.
Emu1 and Emu2 are also shown, which together with the EMILIN/multimerin family have been proposed to form the EDEN superfamily. Red – signal peptide; blue ‘EMI’ – EMI domain; C – coiled-coil region; green ‘COL’ – collagen-like region; pink ‘P’ – proline-rich region; purple ‘EG’ – EGF-like domain.
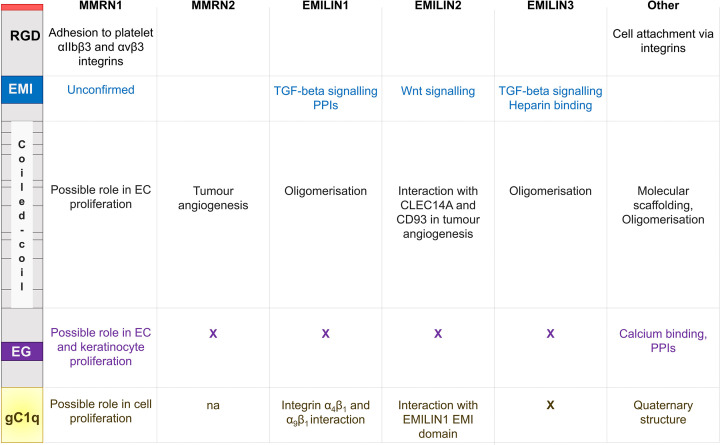
Figure 3. Known and proposed functions of protein domains found in the EMILIN/multimerin family.
The domains and RGD motif of MMRN1 are shown on the left and the reported functions for EMLIN/multimerin family members and other proteins are indicated. Abbreviation: PPI, protein–protein interaction.
Within the EMILIN/multimerin protein family, the EGF-like domain is unique to MMRN1. This domain is associated with calcium binding, mediating protein–protein interactions [53] and is found in proteins linked to cell proliferation, differentiation and cancer, including various ECM proteins [54,55]. An MMRN1-derived peptide corresponding to 11 amino acids of the EGF-like domain has been shown to promote EC and keratinocyte proliferation in vitro [56], but this function is yet to be confirmed for MMRN1 in vivo.
Best known as the globular head domain of the complement protein C1q [57], the gC1q domain plays a role in protein multimerisation and quaternary structure formation [6,58–60]. The EMILIN1 gC1q domain interacts with integrins α4β1 and α9β1, regulating cell adhesion, lymphangiogenesis and tumorigenesis [61–64]. Interestingly, whereas the EMILIN gCq1 domain exerts an anti-proliferative effect, an MMRN1 gC1q-derived peptide has been reported to promote cell proliferation in vitro [56].
Common motifs in protein oligomerisation and macromolecular scaffolding [65], the coiled-coil domain is required for the oligomerisation of EMILIN1 [66] and EMILIN3 [50] and the same role has been inferred for MMRN1 and MMRN2. However, experimental evidence suggests additional roles of the coiled-coil region. Sheets et al. [56] showed that peptides corresponding to MMRN1 coiled-coil regions promoted EC proliferation in vitro. The interaction between the MMRN2 coiled-coil region and CLEC14A and CD93 plays a role in tumour angiogenesis [67].
The N-terminal RGD motif of MMRN1 is common to integrin-interacting ECM proteins and mediates cell adhesion. Integrin specificity, however, depends also on its conformational and spatial presentation [68]. MMRN1 binds to αIIbβ3 and αvβ3 integrins on activated platelets [5], but the interactions of the RGD motif of MMRN1 found in ECs and ECM with integrin has not been confirmed yet. However, adhesive interactions between tumour and ECs form part of the metastasis process and cell–cell interaction may be required for the endothelial transdifferentiation of metastatic melanoma cells to evade detection by the immune system [69]. When analyzing the interaction between melanoma and human umbilical vein endothelial cells (HUVECs), MMRN1 was one of the 30 upregulated genes with a possible role in cell–cell communication and tumour progression [70]. This may suggest a possible mechanism of MMRN1-mediated cell adhesion and melanoma-HUVECs communication. For example, MMRN2’s interaction with CD93 activates β1 integrin signalling, which leads to fibronectin fibrillogenesis and tumour angiogenesis [71]. Considering the importance of integrins in cancer [72], analysis of MMRN1–intergrin interactions in the context of ECs and ECM in cancer may reveal new MMRN1 interaction partners and functions. Such investigations will also confirm the role of proposed MMRN1 protein–protein interactions with serglycin [73,74], which is associated with poor prognosis of disease progression [75], and the oncogene TC2N [76]. The cell proliferative effect of various MMRN1 peptides indicates yet uncharacterised physiological roles of MMRN1 and will need further investigation.
MMRN1 expression profile during development and in healthy tissue
In humans, MMRN1 is considered a marker of early neuroepithelium and is abundantly expressed in long-term self-renewing neuroepithelial-like stem cells [77] and primary osteoblasts [78]. MMRN1 is one of top 20 genes with specific expression in the adult lateral habenula [79]. There also appears to be a sex-specific expression pattern of MMRN1 in human ECs [80].
During mouse development, MMRN1 expression levels are only detectable in differentiated embryonic stem cells, including ECs lining blood vessels (perineural mesenchyme) and mesenchymal cells [81]. Whereas MMRN1 expression levels remain constant in most tissues throughout mouse development and following birth [81], MMRN1 levels increase during the course of murine erythroblast maturation [82]. Although MMRN1 protein levels have been reported to decrease with age [83], RNAseq of murine mammary epithelia and stroma suggests that the proportion and gene expression of lymphatic ECs, for which MMRN1 expression is a marker, remain fairly constant [84].
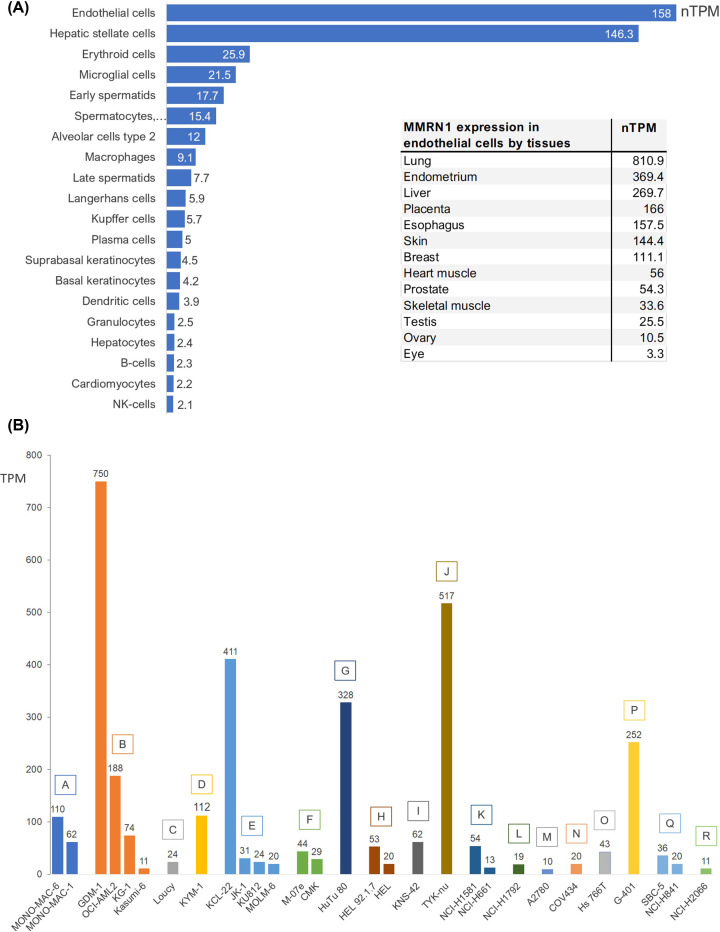
Human MMRN1 is predominantly expressed in ECs, especially in the lung (Figure 4A, data from Human Protein Atlas [85] (http://www.proteinatlas.org/)). It is an established EC marker [86] and mmrn1 has been included in the group of ‘EC-restricted genes’ [87]. ECs show great heterogeneity, providing organ and tissue specific functions, and single-cell RNA sequencing (scRNA) of human lung ECs also identified MMRN1 expression in pulmonary–venous ECs [88]. MMRN1 also shows a distinct expression pattern in the lymphatic EC population in multiple human and murine organs and tissues (heart, muscle, lung, fat, lymph nodes, trachea, liver, middle ear, the eye) [89–95]. Interestingly, single-cell analysis of murine liver cancer identified a new lymphatic EC cluster, using MMRN1 and Pdpn as marker genes [92]. These lymphatic ECs are associated with tumour tissue and have a different gene expression profile to blood vessel ECs, which may suggest a possible relationship with immune cells.
Figure 4. Expression of MMRN1 in cell types and cancer cell lines.
(A) The data show the expression of MMRN1 (nTPM > 2.0) in different healthy cell types. The expression data for these graphs were obtained from the Human Protein Atlas (https://www.proteinatlas.org/ENSG00000138722-MMRN1/single+cell+type) [85]. The inset shows the expression of MMRN1 in epithelial cells in specific organs/tissues. (B) Expression (TPM) of MMRN1 in various cancer cell lines representing adult acute monocytic leukaemia (A), adult acute myeloid leukaemia (B), adult T acute lymphoblastic leukaemia (C), alveolar rhabdomyosarcoma (D), blast phase chronic myelogenous leukaemia (BCR-ABL1 positive, E), childhood acute megakaryoblastic leukaemia (F), duodenal adenocarcinoma (G), erythroleukemia (H), glioblastoma (I), high-grade ovarian serous adenocarcinoma (J), large cell lung carcinoma (K), lung adenocarcinoma (L), ovarian endometrioid adenocarcinoma (M), ovarian granulosa cell tumour (N), pancreatic adenocarcinoma (O), rhabdoid tumour of the kidney (P), small cell lung carcinoma (Q), squamous cell lung carcinoma (R). The expression data were obtained from experiment E-MTAB-2770, EMBL-EBI Expression Atlas (Human Protein Atlas proteinatlas.org, [85]).
MMRN1 differential expression in cancer
Transcriptome analysis of differentially expressed genes (DEGs) aids the identification of hub genes, biomarkers, classification of cancer subtypes and monitoring tumour progression [96–99]. Causes of altered gene expression can include gene mutations, deregulated transcription factors or the action of non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs), which can regulate each other (competitive endogenous RNA (ceRNA) hypothesis) [100,101].
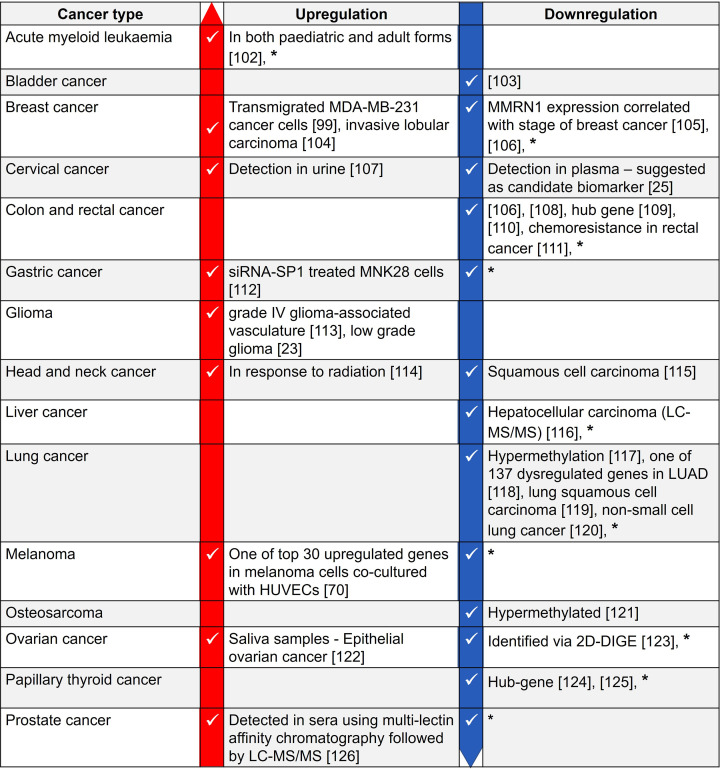
MMRN1 is expressed in various cancer cell lines (Figure 4B), and a DEG in various cancers (Figure 5). It is a potential cancer biomarker in cervical cancer [25] and in paediatric acute myeloid leukaemia [24], where its expression is also positively correlated with the expression of the actin-binding protein Plastin 3 [127]. In rectal cancer, MMRN1 is one of five hub genes that act as prognostic biomarkers and elevated MMRN1 expression is associated with poor prognosis [109]. MMRN1 downregulation is associated with chemoresistance in rectal cancer [111] and radiosensitivity and associated clinical outcome in gastric cancer [128]. The mmrn1 gene is associated with glaucoma [129] and bioinformatics analyses suggest mmrn1 as a hub gene in papillary thyroid cancer [130].
Figure 5. A summary of MMRN1 differential expression in various cancers.
The information is based on RNA-seq and proteomics data published in the stated references and databases. *: MMRN1 differential gene expression as per the TNM plot pan-cancer analysis [47].
In acute myeloid leukaemia, mmrn1 is one of the genes that is upregulated by the action of the oncoprotein MLL-AF6, which is a fusion of the histone methyltransferase mixed lineage leukaemia (MLL) and the cytoplasmic protein AF6 as a result of gene rearrangement [131–133].
MMRN1 expression distinguishes leukaemia stem cells, which contribute to therapy resistance and disease relapse, from leukaemia progenitor cells [134,135]. In adult acute myeloid leukaemia, MMRN1 was identified alongside 16 other genes (AKR1C3, ARHGAP22, CD34, CDK6, CPXM1, EMP1, GPR56, KIAA0125, LAPTM4B, NGFRAP1, NYNRIN, SMIM24, SOCS2, DNMT3B, DPYSL3, ZBTB46), significantly correlated to leukaemia stemness and chemoresistance [102]. This finding led to the 17-gene leukaemia stem cell score (17LSC) which provides a prognostic measure of patient survival [102,136,137]. Similarly, MMRN1 expression levels alongside nine other mRNAs provide a prognostic risk score for gastric cancer patients [138], and Cai et al. [139] included MMRN1 expression in a risk score of papillary thyroid cancer. The observed correlation between MMRN1 expression and cancer risk scores highlights MMRN1 relevance in the disease process. That MMRN1 differential expression is cancer-specific has been shown in head and neck cancer, where MMRN1 downregulation is independent of Human Papilloma virus infections, which is often observed in this cancer type [115]. MMRN1 expression patterns have also been correlated with the stage of breast cancer [105], supporting its potential use in cancer diagnosis [24,139].
Considering the cancer-specific differential expression patterns of MMRN1 and possible application in cancer diagnosis, details on the regulatory mechanisms driving these expression patterns is relatively scarce. Below, we are discussing examples of potential regulatory mechanisms, including methylation, ncRNAs and transcription factors, which have been associated with MMRN1 expression levels in cancers.
MMRN1 regulation via DNA methylation
Modulation of gene expression as a result of DNA methylation is commonly observed in cancer [140]. Although hypermethylation of MMRN1 has been reported in lung adenocarcinoma [117] and osteosarcoma [121], there appears to be no correlation with overall survival in lung cancer or the disease process in osteosarcoma. However, MMRN1 has a possible role in bone remodelling which may be linked to the observation in osteosarcoma [141,142].
MMRN1 regulation via ncRNAs: miRNAs, lncRNAs, circRNAs
The role of miRNAs and lncRNAs in cancer is an ongoing area of research. miRNAs regulate gene expression by suppressing mRNA translation or degrading mRNAs, whereas lncRNAs are molecular scaffolds and form functional complexes with proteins and RNAs to regulate transcription [101,143–145]. ncRNAs are targeting MMRN1 in various cancers (Table 1). Wang et al. [146] showed that an increase in miRNA has-miR-374a (miR-374a), which has known oncogenic properties [147–149], downregulates MMRN1 expression in colorectal adenocarcinoma and colorectal cancer. miR-374a promotes cancer cell proliferation, migration and invasion by downregulating the tumour suppressor gene SRCIN1 (also p140 cas-associated protein) in gastric cancer and regulates Wnt signalling pathways in non-small-cell lung cancer and breast cancer. In colorectal cancer, MMRN1 is targeted by has-miR-99b-5p [143], which is infrequently expressed in overall colorectal tumours, and its upregulation is associated with an increased likelihood of dying.
Table 1. Summary of known ncRNAs regulating MMRN1 in cancers.
| Cancer type | miRNAs | lncRNAs | circRNAs |
|---|---|---|---|
| Colorectal cancer | has-miR-374a has-miR-99b-5p |
||
| Acute myeloid leukaemia | KIAA0125 | ||
| Papillary thyroid cancer | has-miR-4709-3p | LINC00506 | |
| Gastrointestinal stromal tumours | hsa_circ_0070442 |
MMRN1 expression is positively correlated with higher expression of the lncRNA KIAA0125, which, like MMRN1, is included in the prognostic LSC17 for acute myeloid leukaemia [102] and is linked to a poorer prognosis, shorter overall survival and disease-free survival [150].
In papillary thyroid cancer, MMRN1 is co-expressed with the lncRNA LINC00506 but also targeted by the miRNA has-miR-4709-3p. The authors conclude that the interaction LINC00506–MMRN1–has-miR-4709-3p may be specific to this cancer [151].
circRNAs are ncRNAs derived from pre-mRNAs by alternative splicing. Their functions include sponging miRNAs and regulating gene expression and are associated with human diseases including cancer [152]. circRNAs are derived from the genetic information of a so-called ‘host gene’. In gastrointestinal stromal tumours, MMRN1 was identified as the host gene of the circRNA hsa_circ_0070442 and both are downregulated in this cancer. Interestingly, has_circ_0070442 is also one of the top 50 most downregulated circRNAs in lung squamous cell carcinoma [153], a cancer in which MMRN1 expression is also downregulated (based on data in TNM plot [47] and Oncomine [154]). However, a link between has_circ_0070442 and MMRN1 in lung squamous cell carcinoma has not been reported yet.
ceRNAs can be either mRNAs, lncRNAs, circRNAs or even pseudogene gene transcripts, which regulate each other by competitively binding to shared miRNAs [155]. Wen et al. [155] identified TRIB1 with MMRN1 as a ceRNA pair in breast cancer.
Regulation of MMRN1 by ncRNAs is not restricted to disease processes. An integrated transcriptome analysis identified has-miR-514a-3p, which targets MMRN1 in the ovulatory cascade [156].
MMRN1 regulation by transcription factors
Transcription factors control gene expression and their deregulation is associated with cancer [157]. The transcription factor specificity protein 1 (SP1) is overexpressed in many cancers, including gastric cancer, and modulates cell proliferation and survival [158,159]. SP1 regulates MMRN1 expression in vitro in a gastric cancer model. MNK28 cells, which are well-differentiated stomach adenocarcinoma cells, have a high expression of SP1. Silencing SP1 in MNK28 cells with SP1 siRNA results in significant upregulation of MMRN1 (>2.5-fold) [112]. This aligns with the observation that in gastric cancer, where SP1 levels are high, MMRN1 levels are downregulated (Figure 5).
Most information on possible transcription factors that target MMRN1 in various cancers is currently available through bioinformatics analyses and offer an interesting starting point for future in vitro and in vivo experiments. Network analyses identified the transcription factors SP1 and NFIC as regulators of MMRN1 in lung adenocarcinoma [160] and lung squamous cell carcinoma respectively [119]. An association between transcription factor FOX3A and MMRN1 has been reported in anaplastic thyroid carcinoma [161]. The transcription factor-binding sites for ETS-FOXC2 and ETS are present in the mmrn1 gene [162], but there is currently no information if these are targeted to regulate MMRN1 expression in cancers.
MMRN1 alternative splicing and copy number alterations in cancer
The mmrn1 gene is located on chromosome 4 (4q22.1) and contains 11 distinct introns which code for six different isoforms [163]; exon skip events for platelet MMRN1 have been reported previously [164]. Transcriptome analysis of colorectal adenocarcinoma samples identified MMRN1 as one of the 206 genes involved in alternative splicing events in colorectal cancer [165]. In this study, other genes coding for proteins with a role in cell adhesion were identified, but of the original 206 genes, only 18 gene candidates (not including MMRN1) were considered as relevant to cancer development. At the point of writing this review, no other evidence of mmrn1 alternative splicing events in cancer or another disease process was found in the literature.
In non-germinomatous malignant germ cell tumours (NGMGCTs), which are a type of intracranial paediatric germ cell tumour, the mmrn1 gene is deleted due to copy number variations at cytobands 4q13.3-4q28.3 [166]. Studying BIN-67 cells, a model for the rare and aggressive form of small cell ovarian carcinoma of the hypercalcaemic type, SNP array analyses identified copy number loss of 4q22.1, which entails the SNCA and MMRN1 gene [167]. BIN-67 cells are resistant to conventional chemotherapeutics and NGMGCTs are less perceptive to drug and radiation treatment than other germ cell tumours [166]. Although there is no clear involvement of MMRN1 in these cancers on the basis of the studies, it is interesting that these cancers are less responsive to treatment as are other cancers with differential MMRN1 expression e.g., rectal cancer [111].
The effect of signalling cascades and hormones on MMRN1 expression
MMRN1 was classified as an immune-related gene that was only expressed in glioblastoma with high interleukin-13 receptor α2 mRNA expression [168]. Dieterich et al. [113] reported MMRN1 as one of the 78 upregulated genes in the vasculature of grade IV glioma in response to increased VEGF-A and TGFβ2 signalling in the tumour microenvironment. This suggests a possible role of MMRN1 and the other upregulated genes in angiogenesis and VEGFR signalling which are involved in tumorigenesis and metastasis [169].
Hormones like oestrogen and progestogens contribute to breast cancer risk [170,171] and oestrogen can affect gene expression patterns [172,173], including MMRN1. Pitteri et al. [174] reported an increase in serum MMRN1 protein concentration in healthy postmenopausal women who had taken oestrogen and progestin (an exogenous synthetic progestogen) versus oestrogen only. MMRN1 mRNA levels were upregulated in both endometrial stromal cells treated with β-oestradiol [175] and in the endometrium in response to a high serum progesterone level [176]. Whereas MMRN1 levels appear high in these healthy conditions, MMRN1 levels are downregulated in the majority of reported breast cancer datasets (Figure 5). Considering the role of hormones in types of breast cancer, it would be interesting to investigate if MMRN1 is hormonally regulated in the future.
MMRN1 protein detection in cancer
Most of the data of MMRN1 in cancer are based on transcriptional analysis, but MMRN1 protein levels in bodily fluids are being explored in cancer diagnostics Although MMRN1 is considered undetectable in plasma [5], MMRN1 levels have been detected in the plasma, serum, urine and saliva of cancer patients [107,122,123]. Saini et al. [122] report detection of MMRN1 in the saliva of epithelial ovarian cancer patients but comment that the route by which MMRN1 enters the saliva remains unclear. Vizecoumar et al. [177] identified MMRN1 as a downregulated gene whose protein product is detectable in the plasma in gastro-oesophageal cancer patients. Using mass spectrometry techniques, changes in MMRN1 protein levels were detected in the sera of multiple myeloma patients [178] and hepatocellular carcinoma patients [116] and in cell lysates from bone marrow aspirates from patients with amyloid leukaemia [179]. Multi-lectin chromatography, followed by LC-MS/MS identified elevated MMRN1 protein levels in sera from patients with prostate cancer and benign prostate hyperplasia [126]. SWATH-MS identified a positive correlation between MMRN1 and thrombospondin 1 expression, which is differentially regulated in cancers, in the blood plasma of five cancers (colorectal, pancreatic, lung, prostate, ovarian) [180]. Perhaps MMRN1’s presence in various bodily fluids is not entirely a surprise. It has been postulated that proteins are secreted or shed from cancer tissues [180] and it has been shown that MMRN1 is being released via exosomes by duodenal cancer cells [181] (for MMRN1 expression in duodenum adenocarcinoma cell line HuTu80, see Figure 4B), bladder cancer cells [182], and medulloblastoma cells [183]. Exosomes released by UM-SCC6 head-and-neck cancer cells which were treated with ionising radiation had upregulated MMRN1 levels [114]. Exosomes are able to promote metastasis [184], but how MMRN1 exosomal release is associated with cancer progression has not been investigated yet. Elevation of MMRN1 levels in serum exosomes has also been observed in burn patients [185] and patients with tuberculosis infection [186].
Future directions on the functional roles of MMRN1
MMRN1’s potential as a biomarker in certain cancers, including diagnosis of cancer stages, is a strong possibility [23–25,105]. However, detail on MMRN1 protein interactions and involvement in signalling events is required to describe MMRN1 molecular mechanisms and if and how this correlates with its differential expression. If MMRN1 is to be used for cancer diagnosis, detection methods need to be optimised and standardised to provide reliable MMRN1 detection, which can be carried out in a clinical/diagnostic setting e.g., in cervical cancer MMRN1 expression is observed to be either up- or downregulated, depending on whether urine [107] or plasma [25] was analysed (Figure 5). This opposing trend does not exclude MMRN1’s feasibility as biomarker; MMRN1 protein is specifically detected in ovarian cancer [187]. Instead, it highlights the lack of detail to explain MMRN1 protein levels in different bodily fluids and why they vary with disease. For example, MMRN1 has notable mRNA levels in the cervix and cervical mucus, which is a known component of first-void urine samples [188] which may explain MMRN1 detection in urine, but this will need to be verified to support the use MMRN1 as a biomarker in cervical cancer.
Currently, it is difficult to confirm any specific MMRN1 role that may aid or hinder cancer progression, but MMRN1 downregulation in non-small-cell lung cancer has been hypothesised to contribute to vessel leakage and poor blood vessel repair, which would facilitate access of oxygen and nutrients to cancer cells [120]. The in vitro data on MMRN1 peptides corresponding to EGF, gC1q and coiled-coil regions in promoting cell proliferation [56], as well as MMRN1’s potential role in cell adhesion and cell–cell communication [70], certainly warrant further investigations into MMRN1 physiological roles in cancer. Therefore, biochemical and structural studies into MMRN1’s mechanistic roles are needed and may provide support for MMRN1’s use in cancer diagnosis and prognosis.
The role of MMRN1’s cellular localisation is also little understood. Platelets, ECs and the ECM play key roles in cancer. Cross-talk between platelets and tumour cells drives cancer development and progression [41,189] and can facilitate metastasis of solid tumours [40,190]. Malignant tumours also stimulate platelet production (paraneoplastic thrombocytosis), and high platelet numbers correlated with poor cancer prognosis [41,191]. The ECM is remodelled by tumours to create optimal tumorigenic conditions [192]. ECs are embedded in the ECM, both forming part of the tumour microenvironment. Tumour cells regulate ECs to induce processes like angiogenesis, which support their growth and the process of endothelial-to-mesenchymal transition is involved in tumour progression [193]. High-resolution microscopy, proteomics and protein interaction studies can provide information on MMRN1 protein distribution, abundance and interactions in health and disease states and help characterise MMRN1 function in different cellular localisations.
In addition to cancer, MMRN1 is differentially expressed in other diseases including inflammation and bacterial and viral infections [186,194–197]. Upregulation of MMRN1 has also been observed in injuries including septic shock-associated kidney injury [198] and in serum exosomes from burn patients [185]. MMRN1 may also play a role in human pathogen interactions. The MMRN1 protein is the target of Staphylococcus aureus extracellular fibrinogen binding protein (Efb) [199], the Helicobacter pylori vacuolating cytotoxin VacA [200], and MMRN1-derived peptides inhibit Streptococcus pneumoniae adhesion to ECs [201].
The current information indicates that MMRN1 is involved in various disease states that are of medical interest. Transcriptome and bioinformatics analyses have provided the evidence on MMRN1 differential expression. In the next step, characterisation of MMRN1’s physiological functions and molecular mechanisms in ECs and the ECM will be necessary to identify new diagnostic and treatment strategies.
Abbreviations
- ceRNA
competitive endogenous RNA
- circRNA
circular RNA
- DEG
differentially expressed gene
- EC
endothelial cell
- ECM
extracellular matrix
- EGF
epidermal growth factor
- HUVEC
human umbilical vein endothelial cell
- lncRNA
long non-coding RNA
- miRNA
microRNA
- MMRN1
multimerin-1
- ncRNA
non-coding RNA
- NFIC
nuclear factor 1 C-type
- NGMGCT
non-germinomatous malignant germ cell tumour
- RGD motif
arginine-glycine-aspartic acid motif
- SP1
specificity protein 1
- VEGF
vascular endothelial growth factor
Competing Interests
The author declares that there are no competing interests associated with the manuscript.
References
- 1.Hayward C.P., Smith J.W., Horsewood P., Warkentin T.E. and Kelton J.G. (1991) p-155, a multimeric platelet protein that is expressed on activated platelets. J. Biol. Chem. 266, 7114–7120 10.1016/S0021-9258(20)89618-5 [DOI] [PubMed] [Google Scholar]
- 2.Hayward C.P., Bainton D.F., Smith J.W., Horsewood P., Stead R.H., Podor T.J.et al. (1993) Multimerin is found in the alpha-granules of resting platelets and is synthesized by a megakaryocytic cell line. J. Clin. Invest. 91, 2630–2639 10.1172/JCI116502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayward C.P.M., Warkentin T.E., Horsewood P. and Kelton J.G. (1991) Multimerin: a series of large disulfide-linked multimeric proteins within platelets. Blood 77, 2556–2560 10.1182/blood.V77.12.2556.2556 [DOI] [PubMed] [Google Scholar]
- 4.Hayward C.P.M., Hassell J.A., Denomme G.A., Rachubinski R.A., Brown C. and Kelton J.G. (1995) The cDNA Sequence of Human Endothelial Cell Multimerin: a unique protein with RGDs, coiled-coil, and epidermal growth factor-like domains and a carbxyl terminus similar to the globular domain of complement C1q and collagens type VIII and X. J. Biol. Chem. 270, 18246–18251 10.1074/jbc.270.31.18246 [DOI] [PubMed] [Google Scholar]
- 5.Adam F., Zheng S., Joshi N., Kelton D.S., Sandhu A., Suehiro Y.et al. (2005) Analyses of cellular multimerin 1 receptors: in vitro evidence of binding mediated by alphaIIbbeta3 and alphavbeta3. Thromb. Haemost. 94, 1004–1011 10.1160/TH05-02-0140 [DOI] [PubMed] [Google Scholar]
- 6.Colombatti A., Spessotto P., Doliana R., Mongiat M., Bressan G.M. and Esposito G. (2012) The EMILIN/Multimerin family. Front. Immunol. 2, 93– 10.3389/fimmu.2011.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin X., Bern M., Xing Q., Ho J., Viner R. and Mayr M. (2013) Glycoproteomic analysis of the secretome of human endothelial cells*. Mol. Cell. Proteomics 12, 956–978 10.1074/mcp.M112.024018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowenstein C.J., Morrell C.N. and Yamakuchi M. (2005) Regulation of Weibel-Palade body exocytosis. Trends Cardiovasc. Med. 15, 302–308 10.1016/j.tcm.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 9.Heijnen H. and van der Sluijs P. (2015) Platelet secretory behaviour: as diverse as the granules … or not? J. Thromb. Haemost. 13, 2141–2151 10.1111/jth.13147 [DOI] [PubMed] [Google Scholar]
- 10.Tasneem S., Adam F., Minullina I., Pawlikowska M., Hui S.K., Zheng S.et al. (2009) Platelet adhesion to multimerin 1 in vitro: influences of platelet membrane receptors, von Willebrand factor and shear. J. Thromb. Haemost. 7, 685–692 10.1111/j.1538-7836.2009.03284.x [DOI] [PubMed] [Google Scholar]
- 11.Parker D.N., Tasneem S., Farndale R.W., Bihan D., Sadler J.E., Sebastian S.et al. (2016) The functions of the A1A2A3 domains in von Willebrand factor include multimerin 1 binding. Thromb. Haemost. 116, 87–95 10.1160/TH15-09-0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leatherdale A., Parker D.A., Tasneem S., Wang Y., Bihan D., Bonna A.et al. (2021) Multimerin 1 supports platelet function in vivo and binds to specific GPAGPOGPX motifs in fibrillar collagens that enhance platelet adhesion. J. Thromb. Haemost. 19, 547–561 10.1111/jth.15171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leatherdale A., Parker D.A., Tasneem S., Wang Y., Bonna A., Gross P.L.et al. (2017) Multimerin-1 binds to specific motifs in vessel wall collagens and contributes to thrombosis: novel insights regarding the mechanisms that support human and mouse platelet adhesion. Blood 130, 549 [Google Scholar]
- 14.Jeimy S., Woram R., Fuller N., Quinn-Allen M., Nicolaes G., Dahlbäck B.et al. (2004) Identification of the MMRN1 binding region within the C2 domain of human factor V. J. Biol. Chem. 279, 51466–51471 10.1074/jbc.M409866200 [DOI] [PubMed] [Google Scholar]
- 15.Hayward C.P., Fuller N., Zheng S., Adam F., Jeimy S.B., Horsewood I.et al. (2004) Human platelets contain forms of factor V in disulfide-linkage with multimerin. Thromb. Haemost. 92, 1349–1357 10.1160/TH03-02-0123 [DOI] [PubMed] [Google Scholar]
- 16.Jeimy S.B., Fuller N., Tasneem S., Segers K., Stafford A.R., Weitz J.I.et al. (2008) Multimerin 1 binds factor V and activated factor V with high affinity and inhibits thrombin generation. Thromb. Haemost. 100, 1058–1067 10.1160/TH08-05-0307 [DOI] [PubMed] [Google Scholar]
- 17.Jeimy S.B., Krakow E.F., Fuller N., Tasneem S. and Hayward C.P.M. (2008) An acquired factor V inhibitor associated with defective factor V function, storage and binding to multimerin 1. J. Thromb. Haemost. 6, 395–397 10.1111/j.1538-7836.2008.02860.x [DOI] [PubMed] [Google Scholar]
- 18.Jeimy S.B., Quinn-Allen M.A., Fuller N., Kane W.H. and Hayward C.P.M. (2008) Location of the multimerin 1 binding site in coagulation factor V: an update. Thromb. Res. 123, 352–354 10.1016/j.thromres.2008.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veljkovic D.K., Rivard G.E., Diamandis M., Blavignac J., Cramer-Bordé E.M. and Hayward C.P.M. (2009) Increased expression of urokinase plasminogen activator in Quebec platelet disorder is linked to megakaryocyte differentiation. Blood 113, 1535–1542 10.1182/blood-2008-08-172338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunet J.G., Sharma T., Tasneem S., Liang M., Wilson M.D., Rivard G.E.et al. (2020) Thrombin generation abnormalities in Quebec platelet disorder. Int. J. Lab. Hematol. 42, 801–809 10.1111/ijlh.13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahr W.H.A., Zheng S., Sheth P.M., Pai M., Cowie A., Bouchard M.et al. (2001) Platelets from patients with the Quebec platelet disorder contain and secrete abnormal amounts of urokinase-type plasminogen activator. Blood 98, 257–265 10.1182/blood.V98.2.257 [DOI] [PubMed] [Google Scholar]
- 22.Jeimy S.B., Tasneem S., Cramer E.M. and Hayward C.P.M. (2008) Multimerin 1. Platelets 19, 83–95 10.1080/09537100701832157 [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y., Zhang X., Yao J., Jin Z. and Liu C. (2020) Expression patterns and the prognostic value of the EMILIN/Multimerin family members in low-grade glioma. PeerJ 8, e8696–e 10.7717/peerj.8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laszlo G.S., Alonzo T.A., Gudgeon C.J., Harrington K.H., Gerbing R.B., Wang Y.C.et al. (2015) Multimerin-1 (MMRN1) as novel adverse marker in pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. Clin. Cancer Res. 21, 3187–3195 10.1158/1078-0432.CCR-14-2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeratichamroen S., Subhasitanont P., Chokchaichamnankit D., Weeraphan C., Saharat K., Sritana N.et al. (2020) Identification of potential cervical cancer serum biomarkers in Thai patients. Oncol. Lett. 19, 3815–3826 10.3892/ol.2020.11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabajdova M., Urban P., Spakova I., Saksun L., Dudic R., Ostro A.et al. (2016) The crucial role of emilin 1 gene expression during progression of tumor growth. J. Cancer Res. Clin. Oncol. 142, 2397–2402 10.1007/s00432-016-2226-0 [DOI] [PubMed] [Google Scholar]
- 27.Andreuzzi E., Capuano A., Pellicani R., Poletto E., Doliana R., Maiero S.et al. (2018) Loss of Multimerin-2 and EMILIN-2 expression in gastric cancer associate with altered angiogenesis. Int. J. Mol. Sci. 19, 3983 10.3390/ijms19123983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi Y., Lv J., Liu S., Sun L., Wang Y., Li H.et al. (2019) TSPAN9 and EMILIN1 synergistically inhibit the migration and invasion of gastric cancer cells by increasing TSPAN9 expression. BMC Cancer 19, 630 10.1186/s12885-019-5810-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellicani R., Poletto E., Andreuzzi E., Paulitti A., Doliana R., Bizzotto D.et al. (2020) Multimerin-2 maintains vascular stability and permeability. Matrix Biol. 87, 11–25 10.1016/j.matbio.2019.08.002 [DOI] [PubMed] [Google Scholar]
- 30.Mongiat M., Marastoni S., Ligresti G., Lorenzon E., Schiappacassi M., Perris R.et al. (2010) The extracellular matrix glycoprotein elastin microfibril interface located protein 2: a dual role in the tumor microenvironment. Neoplasia 12, 294–304 10.1593/neo.91930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marastoni S., Andreuzzi E., Paulitti A., Colladel R., Pellicani R., Todaro F.et al. (2014) EMILIN2 down-modulates the Wnt signalling pathway and suppresses breast cancer cell growth and migration. J. Pathol. 232, 391–404 10.1002/path.4316 [DOI] [PubMed] [Google Scholar]
- 32.Lorenzon E., Colladel R., Andreuzzi E., Marastoni S., Todaro F., Schiappacassi M.et al. (2012) MULTIMERIN2 impairs tumor angiogenesis and growth by interfering with VEGF-A/VEGFR2 pathway. Oncogene 31, 3136–3147 10.1038/onc.2011.487 [DOI] [PubMed] [Google Scholar]
- 33.Modica T.M.E., Maiorani O., Sartori G., Pivetta E., Doliana R., Capuano A.et al. (2017) The extracellular matrix protein EMILIN1 silences the RAS-ERK pathway via α4β1 integrin and decreases tumor cell growth. Oncotarget 8, 27034–27046 10.18632/oncotarget.15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danussi C., Petrucco A., Wassermann B., Modica T.M.E., Pivetta E., Belluz L.D.B.et al. (2012) An EMILIN1-negative microenvironment promotes tumor cell proliferation and lymph node invasion. Cancer Prev. Res. 5, 1131 10.1158/1940-6207.CAPR-12-0076-T [DOI] [PubMed] [Google Scholar]
- 35.Capuano A., Pivetta E., Sartori G., Bosisio G., Favero A., Cover E.et al. (2019) Abrogation of EMILIN1-β1 integrin interaction promotes experimental colitis and colon carcinogenesis. Matrix Biol. 83, 97–115 10.1016/j.matbio.2019.08.006 [DOI] [PubMed] [Google Scholar]
- 36.Amor López A., Mazariegos M.S., Capuano A., Ximénez-Embún P., Hergueta-Redondo M., Recio J.Á.et al. (2021) Inactivation of EMILIN-1 by proteolysis and secretion in small extracellular vesicles favors melanoma progression and metastasis. Int. J. Mol. Sci. 22, 7406 10.3390/ijms22147406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L.-C., Cui W.-Y., Zhang Z., Tan Z.-L., Lv Q.-L., Chen S.-H.et al. (2021) Expression, methylation and prognostic feature of EMILIN2 in low-grade-glioma. Brain Res. Bull. 175, 26–36 10.1016/j.brainresbull.2021.07.013 [DOI] [PubMed] [Google Scholar]
- 38.Haemmerle M., Stone R.L., Menter D.G., Afshar-Kharghan V. and Sood A.K. (2018) The platelet lifeline to cancer: challenges and opportunities. Cancer Cell 33, 965–983 10.1016/j.ccell.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gkolfinopoulos S., Jones R.L. and Constantinidou A. (2020) The emerging role of platelets in the formation of the micrometastatic niche: current evidence and future perspectives. Front. Oncol. 10, 374 10.3389/fonc.2020.00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucotti S. and Muschel R.J. (2020) Platelets and metastasis: new implications of an old interplay. Front. Oncol. 10, 10.3389/fonc.2020.01350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haemmerle M., Stone R.L., Menter D.G., Afshar-Kharghan V. and Sood A.K. (2018) The platelet lifeline to cancer: challenges and opportunities. Cancer Cell 33, 965–983 10.1016/j.ccell.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hida K., Maishi N., Annan D.A. and Hida Y. (2018) Contribution of tumor endothelial cells in cancer progression. Int. J. Mol. Sci. 19, 1272 10.3390/ijms19051272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopes-Bastos B.M., Jiang W.G. and Cai J. (2016) Tumour–endothelial cell communications: important and indispensable mediators of tumour angiogenesis. Anticancer Res. 36, 1119–1126 [PubMed] [Google Scholar]
- 44.Henke E., Nandigama R. and Ergün S. (2020) Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front. Mol. Biosci. 6, 160 10.3389/fmolb.2019.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cromar G.L., Xiong X., Chautard E., Ricard‐Blum S. and Parkinson J. (2012) Toward a systems level view of the ECM and related proteins: a framework for the systematic definition and analysis of biological systems. Proteins Struct. Funct. Bioinf. 80, 1522–1544 10.1002/prot.24036 [DOI] [PubMed] [Google Scholar]
- 46.Gupta P.B., Pastushenko I., Skibinski A., Blanpain C. and Kuperwasser C. (2019) Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell 24, 65–78 10.1016/j.stem.2018.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartha Á. and Győrffy B. (2021) TNMplot.com: a web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int. J. Mol. Sci. 22, 2622 10.3390/ijms22052622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doliana R., Bot S., Bonaldo P. and Colombatti A. (2000) EMI, a novel cysteine-rich domain of EMILINs and other extracellular proteins, interacts with the gC1q domains and participates in multimerization. FEBS Lett. 484, 164–168 10.1016/S0014-5793(00)02140-2 [DOI] [PubMed] [Google Scholar]
- 49.Zacchigna L., Vecchione C., Notte A., Cordenonsi M., Dupont S., Maretto S.et al. (2006) Emilin1 links TGF-beta maturation to blood pressure homeostasis. Cell 124, 929–942 10.1016/j.cell.2005.12.035 [DOI] [PubMed] [Google Scholar]
- 50.Schiavinato A., Becker A.-K.A., Zanetti M., Corallo D., Milanetto M., Bizzotto D.et al. (2012) EMILIN-3, peculiar member of elastin microfibril interface-located protein (EMILIN) family, has distinct expression pattern, forms oligomeric assemblies, and serves as transforming growth factor β (TGF-β) antagonist. J. Biol. Chem. 287, 11498–11515 10.1074/jbc.M111.303578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bot S., Andreuzzi E., Capuano A., Schiavinato A., Colombatti A. and Doliana R. (2015) Multiple-interactions among EMILIN1 and EMILIN2 N- and C-terminal domains. Matrix Biol. 41, 44–55 10.1016/j.matbio.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 52.Leimeister C., Steidl C., Schumacher N., Erhard S. and Gessler M. (2002) Developmental expression and biochemical characterization of emu family members. Dev. Biol. 249, 204–218 10.1006/dbio.2002.0764 [DOI] [PubMed] [Google Scholar]
- 53.Tombling B.J., Wang C.K. and Craik D.J. (2020) EGF-like and Other disulfide-rich microdomains as therapeutic scaffolds. Angew. Chem. Int. Ed. 59, 11218–11232 10.1002/anie.201913809 [DOI] [PubMed] [Google Scholar]
- 54.Engel J. (1989) EGF-like domains in extracellular matrix proteins: localized signals for growth and differentiation? FEBS Lett. 251, 1–7 10.1016/0014-5793(89)81417-6 [DOI] [PubMed] [Google Scholar]
- 55.Shi S., Ma T. and Xi Y. (2020) A pan-cancer study of epidermal growth factor-like domains 6/7/8 as therapeutic targets in cancer. Front. Genet. 11, 598743 10.3389/fgene.2020.598743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheets A.R., Demidova-Rice T.N., Shi L., Ronfard V., Grover K.V. and Herman I.M. (2016) Identification and characterization of novel matrix-derived bioactive peptides: a role for collagenase from Santyl® ointment in post-debridement wound healing? PLoS ONE 11, e0159598 10.1371/journal.pone.0159598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kishore U., Ghai R., Greenhough T.J., Shrive A.K., Bonifati D.M., Gadjeva M.G.et al. (2004) Structural and functional anatomy of the globular domain of complement protein C1q. Immunol. Lett. 95, 113–128 10.1016/j.imlet.2004.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mongiat M., Mungiguerra G., Bot S., Mucignat M.T., Giacomello E., Doliana R.et al. (2000) Self-assembly and supramolecular organization of EMILIN. J. Biol. Chem. 275, 25471–25480 10.1074/jbc.M001426200 [DOI] [PubMed] [Google Scholar]
- 59.Tang Y.T., Hu T., Arterburn M., Boyle B., Bright J.M., Palencia S.et al. (2005) The complete complement of C1q-domain-containing proteins in Homo sapiens. Genomics 86, 100–111 10.1016/j.ygeno.2005.03.001 [DOI] [PubMed] [Google Scholar]
- 60.Ghebrehiwet B., Hosszu K.K., Valentino A. and Peerschke E.I. (2012) The C1q family of proteins: insights into the emerging non-traditional functions. Front. Immunol. 3, 52 10.3389/fimmu.2012.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capuano A., Pivetta E., Baldissera F., Bosisio G., Wassermann B., Bucciotti F.et al. (2019) Integrin binding site within the gC1q domain orchestrates EMILIN-1-induced lymphangiogenesis. Matrix Biol. 81, 34–49 10.1016/j.matbio.2018.10.006 [DOI] [PubMed] [Google Scholar]
- 62.Spessotto P., Cervi M., Mucignat M.T., Mungiguerra G., Sartoretto I., Doliana R.et al. (2003) β1 integrin-dependent cell adhesion to EMILIN-1 is mediated by the gC1q domain*. J. Biol. Chem. 278, 6160–6167 10.1074/jbc.M208322200 [DOI] [PubMed] [Google Scholar]
- 63.Verdone G., Doliana R., Corazza A., Colebrooke S.A., Spessotto P., Bot S.et al. (2008) The solution structure of EMILIN1 globular C1q domain reveals a disordered insertion necessary for interaction with the α4β1 integrin*. J. Biol. Chem. 283, 18947–18956 10.1074/jbc.M801085200 [DOI] [PubMed] [Google Scholar]
- 64.Maiorani O., Pivetta E., Capuano A., Modica T.M.E., Wassermann B., Bucciotti F.et al. (2017) Neutrophil elastase cleavage of the gC1q domain impairs the EMILIN1-α4β1 integrin interaction, cell adhesion and anti-proliferative activity. Sci. Rep. 7, 39974– 10.1038/srep39974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McAlinden A., Smith T.A., Sandell L.J., Ficheux D., Parry D.A.D. and Hulmes D.J.S. (2003) α-helical coiled-coil oligomerization domains are almost ubiquitous in the collagen superfamily*. J. Biol. Chem. 278, 42200–42207 10.1074/jbc.M302429200 [DOI] [PubMed] [Google Scholar]
- 66.Iacomino M., Doliana R., Marchese M., Capuano A., Striano P., Spessotto P.et al. (2020) Distal motor neuropathy associated with novel EMILIN1 mutation. Neurobiol. Dis. 137, 104757 10.1016/j.nbd.2020.104757 [DOI] [PubMed] [Google Scholar]
- 67.Khan K.A., Naylor A.J., Khan A., Noy P.J., Mambretti M., Lodhia P.et al. (2017) Multimerin-2 is a ligand for group 14 family C-type lectins CLEC14A, CD93 and CD248 spanning the endothelial pericyte interface. Oncogene 36, 6097–6108 10.1038/onc.2017.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nieberler M., Reuning U., Reichart F., Notni J., Wester H.-J., Schwaiger M.et al. (2017) Exploring the role of RGD-recognizing integrins in cancer. Cancers 9, 116 10.3390/cancers9090116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X., Karras P., Torres R., Rambow F., van den Oord J., Marine J.-C.et al. (2020) Disseminated melanoma cells transdifferentiate into endothelial cells in intravascular niches at metastatic sites. Cell Rep. 31, 107765 10.1016/j.celrep.2020.107765 [DOI] [PubMed] [Google Scholar]
- 70.Stine M.J., Wang C.J., Moriarty W.F., Ryu B., Cheong R., Westra W.H.et al. (2011) Integration of genotypic and phenotypic screening reveals molecular mediators of melanoma-stromal interaction. Cancer Res. 71, 2433–2444 10.1158/0008-5472.CAN-10-1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lugano R., Vemuri K., Yu D., Bergqvist M., Smits A., Essand M.et al. (2018) CD93 promotes β1 integrin activation and fibronectin fibrillogenesis during tumor angiogenesis. J. Clin. Invest. 128, 3280–3297 10.1172/JCI97459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desgrosellier J.S. and Cheresh D.A. (2010) Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22 10.1038/nrc2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Korpetinou A., Skandalis S.S., Labropoulou V.T., Smirlaki G., Noulas A., Karamanos N.K.et al. (2014) Serglycin: at the crossroad of inflammation and malignancy. Front. Oncol. 3, 327 10.3389/fonc.2013.00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manou D., Karamanos N.K. and Theocharis A.D. (2020) Tumorigenic functions of serglycin: Regulatory roles in epithelial to mesenchymal transition and oncogenic signaling. Semin. Cancer Biol. 62, 108–115 10.1016/j.semcancer.2019.07.004 [DOI] [PubMed] [Google Scholar]
- 75.Guo J.-Y., Chiu C.-H., Wang M.-J., Li F.-A. and Chen J.-Y. (2020) Proteoglycan serglycin promotes non-small cell lung cancer cell migration through the interaction of its glycosaminoglycans with CD44. J. Biomed. Sci. 27, 2 10.1186/s12929-019-0600-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qureshi M.A., Khan S., Tauheed M.S., Syed S.A., Ujjan I.D., Lail A.et al. (2020) Pan-cancer multiomics analysis of TC2N gene suggests its important role(s) in tumourigenesis of many cancers. Asian Pac. J. Cancer Prev. 21, 3199–3209 10.31557/APJCP.2020.21.11.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Falk A., Koch P., Kesavan J., Takashima Y., Ladewig J., Alexander M.et al. (2012) Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS ONE 7, e29597 10.1371/journal.pone.0029597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hsu Y.-H., Zillikens M.C., Wilson S.G., Farber C.R., Demissie S., Soranzo N.et al. (2010) An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility loci for osteoporosis-related traits. PLoS Genet. 6, e1000977 10.1371/journal.pgen.1000977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Foll B. and French L. (2018) Transcriptomic characterization of the human Habenula highlights drug metabolism and the neuroimmune system. Front. Neurosci. 12, 742 10.3389/fnins.2018.00742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Witt E., Lorenz M., Völker U., Stangl K., Hammer E. and Stangl V. (2019) Sex-specific differences in the intracellular proteome of human endothelial cells from dizygotic twins. J. Proteomics 201, 48–56 10.1016/j.jprot.2019.03.016 [DOI] [PubMed] [Google Scholar]
- 81.Braghetta P., Ferrari A., de Gemmis P., Zanetti M., Volpin D., Bonaldo P.et al. (2004) Overlapping, complementary and site-specific expression pattern of genes of the EMILIN/Multimerin family. Matrix Biol. 22, 549–556 10.1016/j.matbio.2003.10.005 [DOI] [PubMed] [Google Scholar]
- 82.Yu X., Martella A., Kolovos P., Stevens M., Stadhouders R., Grosveld F.G.et al. (2020) The dynamic emergence of GATA1 complexes identified in in vitro embryonic stem cell differentiation and in vivo mouse fetal liver. Haematologica 105, 1802–1812 10.3324/haematol.2019.216010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang H., Zhu X., Shen J., Zhao E.-F., He D., Shen H.et al. (2019) Quantitative iTRAQ-based proteomic analysis of differentially expressed proteins in aging in human and monkey. BMC Genomics 20, 725 10.1186/s12864-019-6089-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li C.M.-C., Shapiro H., Tsiobikas C., Selfors L.M., Chen H., Rosenbluth J.et al. (2020) Aging-associated alterations in mammary epithelia and stroma revealed by single-cell RNA sequencing. Cell Rep. 33, 108566 10.1016/j.celrep.2020.108566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A.et al. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 86.Wessells H., Sullivan C.J., Tsubota Y., Engel K.L., Kim B., Olson N.E.et al. (2009) Transcriptional profiling of human cavernosal endothelial cells reveals distinctive cell adhesion phenotype and role for claudin 11 in vascular barrier function. Physiol. Genomics 39, 100–108 10.1152/physiolgenomics.90354.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhasin M., Yuan L., Keskin D.B., Otu H.H., Libermann T.A. and Oettgen P. (2010) Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genomics 11, 342 10.1186/1471-2164-11-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schupp J.C., Adams T.S., Cosme C., Raredon M.S.B., Omote N., De Frias S.P.et al. (2021) Integrated single cell atlas of endothelial cells of the human lung. Circulation 144, 286–302 10.1161/CIRCULATIONAHA.120.052318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L., Yang Y., Ma H., Xie Y., Xu J., Near D.et al. (2021) Single cell dual-omics reveals the transcriptomic and epigenomic diversity of cardiac non-myocytes. Cardiovasc. Res. cvab134 10.1093/cvr/cvab134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng W., Chen L., Nguyen P.K., Wu S.M. and Li G. (2019) Single cell analysis of endothelial cells identified organ-specific molecular signatures and heart-specific cell populations and molecular features. Front. Cardiovasc. Med. 6, 165 10.3389/fcvm.2019.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao Q., Eichten A., Parveen A., Adler C., Huang Y., Wang W.et al. (2018) Single-cell transcriptome analyses reveal endothelial cell heterogeneity in tumors and changes following antiangiogenic treatment. Cancer Res. 78, 2370–2382 10.1158/0008-5472.CAN-17-2728 [DOI] [PubMed] [Google Scholar]
- 92.Zhao Q., Molina-Portela M.D.P., Parveen A., Adler A., Adler C., Hock E.et al. (2020) Heterogeneity and chimerism of endothelial cells revealed by single-cell transcriptome in orthotopic liver tumors. Angiogenesis 23, 581–597 10.1007/s10456-020-09727-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ryan A.F., Nasamran C.A., Pak K., Draf C., Fisch K.M., Webster N.et al. (2020) Single-cell transcriptomes reveal a complex cellular landscape in the middle ear and differential capacities for acute response to infection. Front. Genet. 11, 10.3389/fgene.2020.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patel G., Fury W., Yang H., Gomez-Caraballo M., Bai Y., Yang T.et al. (2020) Molecular taxonomy of human ocular outflow tissues defined by single-cell transcriptomics. Proc. Natl. Acad. Sci. U.S.A. 117, 12856–12867 10.1073/pnas.2001896117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xiang M., Grosso R.A., Takeda A., Pan J., Bekkhus T., Brulois K.et al. (2020) A single-cell transcriptional roadmap of the mouse and human lymph node lymphatic vasculature. Front. Cardiovasc. Med. 7, 52 10.3389/fcvm.2020.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xue J.-M., Liu Y., Wan L.-H. and Zhu Y.-X. (2020) Comprehensive analysis of differential gene expression to identify common gene signatures in multiple cancers. Med. Sci. Monit. 26, e919953–e 10.12659/MSM.919953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee G. and Lee M. (2017) Classification of genes based on age-related differential expression in breast cancer. Genomics Inform. 15, 156–161 10.5808/GI.2017.15.4.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rosario S.R., Long M.D., Affronti H.C., Rowsam A.M., Eng K.H. and Smiraglia D.J. (2018) Pan-cancer analysis of transcriptional metabolic dysregulation using The Cancer Genome Atlas. Nat. Commun. 9, 5330 10.1038/s41467-018-07232-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bersini S., Miermont A., Pavesi A., Kamm R.D., Thiery J.P., Moretti M.et al. (2018) A combined microfluidic-transcriptomic approach to characterize the extravasation potential of cancer cells. Oncotarget 9, 36110–36125 10.18632/oncotarget.26306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cai Y. and Wan J. (2018) Competing endogenous RNA regulations in neurodegenerative disorders: current challenges and emerging insights. Front. Mol. Neurosci. 11, 370– 10.3389/fnmol.2018.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin C.-P. and He L. (2017) Noncoding RNAs in cancer development. Ann. Rev. Cancer Biol. 1, 163–184 10.1146/annurev-cancerbio-050216-034443 [DOI] [Google Scholar]
- 102.Ng S.W.K., Mitchell A., Kennedy J.A., Chen W.C., McLeod J., Ibrahimova N.et al. (2016) A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 540, 433–437 10.1038/nature20598 [DOI] [PubMed] [Google Scholar]
- 103.Ma Y., Feng X.-F., Yang W.-X. and You C.-G. (2019) Exploring the pathological mechanism of bladder cancer based on tumor mutational burden analysis. Biomed Res. Int. 2019, 1093815 10.1155/2019/1093815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao H., Langerød A., Ji Y., Nowels K.W., Nesland J.M., Tibshirani R.et al. (2004) Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol. Biol. Cell 15, 2523–2536 10.1091/mbc.e03-11-0786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shi Y., Steppi A., Cao Y., Wang J., He M.M., Li L.et al. (2017) Integrative comparison of mRNA expression patterns in breast cancers from Caucasian and Asian Americans with implications for precision medicine. Cancer Res. 77, 423–433 10.1158/0008-5472.CAN-16-1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rachidi S.M., Qin T., Sun S., Zheng W.J. and Li Z. (2013) Molecular profiling of multiple human cancers defines an inflammatory cancer-associated molecular pattern and uncovers KPNA2 as a uniform poor prognostic cancer marker. PLoS ONE 8, e57911–e 10.1371/journal.pone.0057911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chokchaichamnankit D., Watcharatanyatip K., Subhasitanont P., Weeraphan C., Keeratichamroen S., Sritana N.et al. (2019) Urinary biomarkers for the diagnosis of cervical cancer by quantitative label-free mass spectrometry analysis. Oncol. Lett. 17, 5453–5468 10.3892/ol.2019.10227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang W., Ma J., Zhou W., Li Z., Zhou X., Cao B.et al. (2019) Identification of hub genes and outcome in colon cancer based on bioinformatics analysis. Cancer Manag. Res. 11, 323–338 10.2147/CMAR.S173240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li C., Liu T., Liu Y., Zhang J. and Zuo D. (2021) Prognostic value of tumour microenvironment‐related genes by TCGA database in rectal cancer. J. Cell. Mol. Med. 25, 5811– 5822 10.1111/jcmm.16547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun F., Liang W., Tang K., Hong M. and Qian J. (2019) Profiling the lncRNA-miRNA-mRNA ceRNA network to reveal potential crosstalk between inflammatory bowel disease and colorectal cancer. PeerJ 7, e7451–e 10.7717/peerj.7451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gantt G.A., Chen Y., DeJulius K., Mace A.G., Barnholtz-Sloan J. and Kalady M.F. (2014) Gene expression profile is associated with chemoradiation resistance in rectal cancer. Colorectal Dis. 16, 57–66 10.1111/codi.12395 [DOI] [PubMed] [Google Scholar]
- 112.Lee H.S., Park C.-K., Oh E., Erkin Ö.C., Jung H.S., Cho M.-H.et al. (2013) Low SP1 expression differentially affects intestinal-type compared with diffuse-type gastric adenocarcinoma. PLoS ONE 8, e55522 10.1371/journal.pone.0055522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dieterich L.C., Mellberg S., Langenkamp E., Zhang L., Zieba A., Salomäki H.et al. (2012) Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF‐A and TGFβ2 in vascular abnormalization. J. Pathol. 228, 378–390 10.1002/path.4072 [DOI] [PubMed] [Google Scholar]
- 114.Abramowicz A., Wojakowska A., Marczak L., Lysek-Gladysinska M., Smolarz M., Story M.D.et al. (2019) Ionizing radiation affects the composition of the proteome of extracellular vesicles released by head-and-neck cancer cells in vitro. J. Radiat. Res. 60, rrz001 10.1093/jrr/rrz001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martinez I., Wang J., Hobson K.F., Ferris R.L. and Khan S.A. (2007) Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur. J. Cancer 43, 415–432 10.1016/j.ejca.2006.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tsai T.-H., Song E., Zhu R., Di Poto C., Wang M., Luo Y.et al. (2015) LC-MS/MS-based serum proteomics for identification of candidate biomarkers for hepatocellular carcinoma. Proteomics 15, 2369–2381 10.1002/pmic.201400364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu X.-F., Zhu B.-S., Wu F.-M. and Hu H.-B. (2018) DNA methylation biomarkers for the occurrence of lung adenocarcinoma from TCGA data mining. J. Cell. Physiol. 233, 6777–6784 10.1002/jcp.26531 [DOI] [PubMed] [Google Scholar]
- 118.Wang Y., Zhou Z., Chen L., Li Y., Zhou Z. and Chu X. (2021) Identification of key genes and biological pathways in lung adenocarcinoma via bioinformatics analysis. Mol. Cell. Biochem. 476, 931–939 10.1007/s11010-020-03959-5 [DOI] [PubMed] [Google Scholar]
- 119.Zhang F., Chen X., Wei K., Liu D., Xu X., Zhang X.et al. (2017) Identification of key transcription factors associated with lung squamous cell carcinoma. Med. Sci. Monit. 23, 172–206 10.12659/MSM.898297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Välk K., Vooder T., Kolde R., Reintam M.A., Petzold C., Vilo J.et al. (2010) Gene expression profiles of non-small cell lung cancer: survival prediction and new biomarkers. Oncology 79, 283–292 10.1159/000322116 [DOI] [PubMed] [Google Scholar]
- 121.Wang T.-X., Tan W.-L., Huang J.-C., Cui Z.-F., Liang R.-D., Li Q.-C.et al. (2020) Identification of aberrantly methylated differentially expressed genes targeted by differentially expressed miRNA in osteosarcoma. Ann. Transl. Med. 8, 373 10.21037/atm.2020.02.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saini A., Chandra K.B., Kumar V., Mathur S.R., Sharma J.B., Kumar S.et al. (2020) Analysis of Multimerin 1 (MMRN1) expression in ovarian cancer. Mol. Biol. Rep. 47, 9459–9468 10.1007/s11033-020-06027-9 [DOI] [PubMed] [Google Scholar]
- 123.Tajmul M., Parween F., Singh L., Mathur S.R., Sharma J.B., Kumar S.et al. (2018) Identification and validation of salivary proteomic signatures for non-invasive detection of ovarian cancer. Int. J. Biol. Macromol. 108, 503–514 10.1016/j.ijbiomac.2017.12.014 [DOI] [PubMed] [Google Scholar]
- 124.Zhang K., Liu J., Li C., Peng X., Li H. and Li Z. (2019) Identification and validation of potential target genes in papillary thyroid cancer. Eur. J. Pharmacol. 843, 217–225 10.1016/j.ejphar.2018.11.026 [DOI] [PubMed] [Google Scholar]
- 125.Wang Q., Shen Y., Ye B., Hu H., Fan C., Wang T.et al. (2018) Gene expression differences between thyroid carcinoma, thyroid adenoma and normal thyroid tissue. Oncol. Rep. 40, 3359–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Totten S.M., Adusumilli R., Kullolli M., Tanimoto C., Brooks J.D., Mallick P.et al. (2018) Multi-lectin affinity chromatography and quantitative proteomic analysis reveal differential glycoform levels between prostate cancer and benign prostatic hyperplasia sera. Sci. Rep. 8, 6509 10.1038/s41598-018-24270-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Velthaus A., Cornils K., Hennigs J.K., Grüb S., Stamm H., Wicklein D.et al. (2019) The actin binding protein Plastin-3 Is involved in the pathogenesis of acute myeloid leukemia. Cancers 11, 1663 10.3390/cancers11111663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liang L.-B., Huang X.-Y., He H. and Liu J.-Y. (2020) Prognostic values of radiosensitivity genes and CD19 status in gastric cancer: a retrospective study using TCGA database. Pharmgenomics Pers. Med. 13, 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moazzeni H., Khani M. and Elahi E. (2020) Insights into the regulatory molecules involved in glaucoma pathogenesis. Am. J. Med. Genet. C Semin. Med. Genet. 184, 782–827 10.1002/ajmg.c.31833 [DOI] [PubMed] [Google Scholar]
- 130.Zhang K., Liu J., Li C., Peng X., Li H., and Li Z. (2019) Identification and validation of potential target genes in papillary thyroid cancer. Eur. J. Pharmacol. 15, 217–225 10.1016/j.ejphar.2018.11.026 [DOI] [PubMed] [Google Scholar]
- 131.Deshpande A.J., Chen L., Fazio M., Sinha A.U., Bernt K.M., Banka D.et al. (2013) Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l. Blood 121, 2533–2541 10.1182/blood-2012-11-465120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Numata A., Kwok H.S., Kawasaki A., Li J., Zhou Q.-L., Kerry J.et al. (2018) The basic helix-loop-helix transcription factor SHARP1 is an oncogenic driver in MLL-AF6 acute myelogenous leukemia. Nat. Commun. 9, 1622 10.1038/s41467-018-03854-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Manara E., Baron E., Tregnago C., Aveic S., Bisio V., Bresolin S.et al. (2014) MLL-AF6 fusion oncogene sequesters AF6 into the nucleus to trigger RAS activation in myeloid leukemia. Blood 124, 263–272 10.1182/blood-2013-09-525741 [DOI] [PubMed] [Google Scholar]
- 134.Gentles A.J., Plevritis S.K., Majeti R. and Alizadeh A.A. (2010) Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA 304, 2706–2715 10.1001/jama.2010.1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arnone M., Konantz M., Hanns P., Paczulla Stanger A.M., Bertels S., Godavarthy P.S.et al. (2020) Acute myeloid leukemia stem cells: the challenges of phenotypic heterogeneity. Cancers (Basel) 12, 3742 10.3390/cancers12123742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang F., Morita K., DiNardo C.D., Furudate K., Tanaka T., Yan Y.et al. (2021) Leukemia stemness and co-occurring mutations drive resistance to IDH inhibitors in acute myeloid leukemia. Nat. Commun. 12, 2607 10.1038/s41467-021-22874-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bill M., Nicolet D., Kohlschmidt J., Walker C.J., Mrózek K., Eisfeld A.-K.et al. (2020) Mutations associated with a 17-gene leukemia stem cell score and its prognostic relevance in the context of the European LeukemiaNet classification for acute myeloid leukemia. Haematologica 105, 721–729haematol.2019.225003 10.3324/haematol.2019.225003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sun M., Qiu J., Zhai H., Wang Y., Ma P., Li M.et al. (2020) Prognostic implications of novel gene signatures in gastric cancer microenvironment. Med. Sci. Monit. 26, e924604 10.12659/MSM.924604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cai W.Y., Chen X., Chen L.P., Li Q., Du X.J. and Zhou Y.Y. (2018) Role of differentially expressed genes and long non‐coding RNAs in papillary thyroid carcinoma diagnosis, progression, and prognosis. J. Cell. Biochem. 119, 8249–8259 10.1002/jcb.26836 [DOI] [PubMed] [Google Scholar]
- 140.Skvortsova K., Stirzaker C. and Taberlay P. (2019) The DNA methylation landscape in cancer. Essays Biochem. 63, 797–811 10.1042/EBC20190037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liron T., Raphael B., Hiram-Bab S., Bab I.A. and Gabet Y. (2018) Bone loss in C57BL/6J-OlaHsd mice, a substrain of C57BL/6J carrying mutated alpha-synuclein and multimerin-1 genes. J. Cell. Physiol. 233, 371–377 10.1002/jcp.25895 [DOI] [PubMed] [Google Scholar]
- 142.Huang D., Liu J., Cao Y., Wan L., Jiang H., Sun Y.et al. (2020) RNA sequencing for gene expression profiles in peripheral blood mononuclear cells with ankylosing spondylitis RNA. Biomed Res. Int. 2020, 5304578 10.1155/2020/5304578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Slattery M.L., Pellatt A.J., Lee F.Y., Herrick J.S., Samowitz W.S., Stevens J.R.et al. (2017) Infrequently expressed miRNAs influence survival after diagnosis with colorectal cancer. Oncotarget 8, 83845–83859 10.18632/oncotarget.19863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guttman M. and Rinn J.L. (2012) Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang G., Lu X. and Yuan L. (2014) LncRNA: a link between RNA and cancer. Biochim. Biophys. Acta Gene Regul. Mech. 1839, 1097–1109 [DOI] [PubMed] [Google Scholar]
- 146.Wang Y., Zhang J., Li L., Xu X., Zhang Y., Teng Z.et al. (2016) Identification of molecular targets for predicting colon adenocarcinoma. Med. Sci. Monit. 22, 460–468 10.12659/MSM.895881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu X., Wang W., Su N., Zhu X., Yao J., Gao W.et al. (2015) miR-374a promotes cell proliferation, migration and invasion by targeting SRCIN1 in gastric cancer. FEBS Lett. 589, 407–413 10.1016/j.febslet.2014.12.027 [DOI] [PubMed] [Google Scholar]
- 148.Wang Y., Xia H., Zhuang Z., Miao L., Chen X. and Cai H. (2014) Axl-altered microRNAs regulate tumorigenicity and gefitinib resistance in lung cancer. Cell Death Dis. 5, e1227–e 10.1038/cddis.2014.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cai J., Guan H., Fang L., Yang Y., Zhu X., Yuan J.et al. (2013) MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J. Clin. Invest. 123, 566–579 10.1172/JCI65871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang Y.H., Lin C.C., Hsu C.L., Hung S.Y., Yao C.Y., Lee S.H.et al. (2021) Distinct clinical and biological characteristics of acute myeloid leukemia with higher expression of long noncoding RNA KIAA0125. Ann. Hematol. 100, 487–498 10.1007/s00277-020-04358-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang F., Zhang J., Li B., Zhao Z., Liu Y., Zhao Z.et al. (2021) Identification of potential lncRNAs and miRNAs as diagnostic biomarkers for papillary thyroid carcinoma based on machine learning. Int. J. Endocrinol. 2021, 3984463– 10.1155/2021/3984463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Su M., Xiao Y., Ma J., Tang Y., Tian B., Zhang Y.et al. (2019) Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 18, 90 10.1186/s12943-019-1002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cheng Z., Yu C., Cui S., Wang H., Jin H., Wang C.et al. (2019) circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat. Commun. 10, 3200 10.1038/s41467-019-11162-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D.et al. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wen X., Gao L. and Hu Y. (2020) LAceModule: identification of competing endogenous RNA modules by integrating dynamic correlation. Front. Genet. 11, 235 10.3389/fgene.2020.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yerushalmi G.M., Salmon-Divon M., Ophir L., Yung Y., Baum M., Coticchio G.et al. (2018) Characterization of the miRNA regulators of the human ovulatory cascade. Sci. Rep. 8, 15605 10.1038/s41598-018-33807-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M.et al. (2018) The human transcription factors. Cell 172, 650–665 10.1016/j.cell.2018.01.029 [DOI] [PubMed] [Google Scholar]
- 158.Beishline K. and Azizkhan-Clifford J. (2015) Sp1 and the ‘hallmarks of cancer’. FEBS J. 282, 224–258 10.1111/febs.13148 [DOI] [PubMed] [Google Scholar]
- 159.Deniaud E., Baguet J., Mathieu A.L., Pagès G., Marvel J. and Leverrier Y. (2006) Overexpression of Sp1 transcription factor induces apoptosis. Oncogene 25, 7096–7105 10.1038/sj.onc.1209696 [DOI] [PubMed] [Google Scholar]
- 160.Li J., Li Z., Zhao S., Song Y., Si L. and Wang X. (2020) Identification key genes, key miRNAs and key transcription factors of lung adenocarcinoma. J. Thorac. Dis. 12, 1917–1933 10.21037/jtd-19-4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pan Z., Li L., Fang Q., Qian Y., Zhang Y., Zhu J.et al. (2019) Integrated bioinformatics analysis of master regulators in anaplastic thyroid carcinoma. Biomed Res. Int. 2019, 9734576 10.1155/2019/9734576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Miano J.M. and Long X. (2015) The short and long of noncoding sequences in the control of vascular cell phenotypes. Cell. Mol. Life Sci. 72, 3457–3488 10.1007/s00018-015-1936-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Thierry-Mieg D. and Thierry-Mieg J. (2006) AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 7, S12 10.1186/gb-2006-7-s1-s12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Power K.A., McRedmond J.P., de Stefani A., Gallagher W.M. and Ó Gaora P. (2009) High-throughput proteomics detection of novel splice isoforms in human platelets. PLoS ONE 4, e5001 10.1371/journal.pone.0005001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bisognin A., Pizzini S., Perilli L., Esposito G., Mocellin S., Nitti D.et al. (2014) An integrative framework identifies alternative splicing events in colorectal cancer development. Mol. Oncol. 8, 129–141 10.1016/j.molonc.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang H.-W., Wu Y.-H., Hsieh J.-Y., Liang M.-L., Chao M.-E., Liu D.-J.et al. (2010) Pediatric primary central nervous system germ cell tumors of different prognosis groups show characteristic miRNome traits and chromosome copy number variations. BMC Genomics 11, 132– 10.1186/1471-2164-11-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gamwell L.F., Gambaro K., Merziotis M., Crane C., Arcand S.L., Bourada V.et al. (2013) Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics. Orphanet J. Rare Dis. 8, 33 10.1186/1750-1172-8-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Han J. and Puri R.K. (2018) Analysis of the cancer genome atlas (TCGA) database identifies an inverse relationship between interleukin-13 receptor α1 and α2 gene expression and poor prognosis and drug resistance in subjects with glioblastoma multiforme. J. Neuro Oncol. 136, 463–474 10.1007/s11060-017-2680-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nishida N., Yano H., Nishida T., Kamura T. and Kojiro M. (2006) Angiogenesis in cancer. Vasc. Health Risk Manag. 2, 213–219 10.2147/vhrm.2006.2.3.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Trabert B., Sherman M.E., Kannan N. and Stanczyk F.Z. (2020) Progesterone and breast cancer. Endocr. Rev. 41, 320–344 10.1210/endrev/bnz001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Travis R.C. and Key T.J. (2003) Oestrogen exposure and breast cancer risk. Breast Cancer Res. 5, 239–247 10.1186/bcr628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Frasor J., Danes J.M., Komm B., Chang K.C.N., Lyttle C.R. and Katzenellenbogen B.S. (2003) Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144, 4562–4574 10.1210/en.2003-0567 [DOI] [PubMed] [Google Scholar]
- 173.Murphy E. and Steenbergen C. (2014) Estrogen regulation of protein expression and signaling pathways in the heart. Biol. Sex Differ. 5, 6– 10.1186/2042-6410-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pitteri S.J., Hanash S.M., Aragaki A., Amon L.M., Chen L., Busald Buson T.et al. (2009) Postmenopausal estrogen and progestin effects on the serum proteome. Genome Med. 1, 121 10.1186/gm121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ren C.-E., Zhu X., Li J., Lyle C., Dowdy S., Podratz K.C.et al. (2015) Microarray analysis on gene regulation by estrogen, progesterone and tamoxifen in human endometrial stromal cells. Int. J. Mol. Sci. 16, 5864–5885 10.3390/ijms16035864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Labarta E., Martínez-Conejero J.A., Alamá P., Horcajadas J.A., Pellicer A., Simón C.et al. (2011) Endometrial receptivity is affected in women with high circulating progesterone levels at the end of the follicular phase: a functional genomics analysis. Hum. Reprod. 26, 1813–1825 10.1093/humrep/der126 [DOI] [PubMed] [Google Scholar]
- 177.Vizeacoumar F.S., Guo H., Dwernychuk L., Zaidi A., Freywald A., Wu F.-X.et al. (2021) Mining the plasma-proteome associated genes in patients with gastro-esophageal cancers for biomarker discovery. Sci. Rep. 11, 7590 10.1038/s41598-021-87037-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang H.-T., Tian E.-B., Chen Y.-L., Deng H.-T. and Wang Q.-T. (2015) Proteomic analysis for finding serum pathogenic factors and potential biomarkers in multiple myeloma. Chin. Med. J. (Engl.) 128, 1108–1113 10.4103/0366-6999.155112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tong J., Helmy M., Cavalli F.M., Jin L., St-Germain J., Karisch R.et al. (2017) Integrated analysis of proteome, phosphotyrosine-proteome, tyrosine-kinome, and tyrosine-phosphatome in acute myeloid leukemia. Proteomics 17, 10.1002/pmic.201600361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sajic T., Liu Y., Arvaniti E., Surinova S., Williams E.G., Schiess R.et al. (2018) Similarities and differences of blood N-glycoproteins in five solid carcinomas at localized clinical stage analyzed by SWATH-MS. Cell Rep. 23, 2819.e5–2831.e5 10.1016/j.celrep.2018.04.114 [DOI] [PubMed] [Google Scholar]
- 181.Ohshima K., Kanto K., Hatakeyama K., Ide T., Wakabayashi-Nakao K., Watanabe Y.et al. (2014) Exosome-mediated extracellular release of polyadenylate-binding protein 1 in human metastatic duodenal cancer cells. Proteomics 14, 2297–2306 10.1002/pmic.201300477 [DOI] [PubMed] [Google Scholar]
- 182.Kumari N., Saxena S. and Agrawal U. (2015) Exosomal protein interactors as emerging therapeutic targets in urothelial bladder cancer. J. Egypt. Natl. Cancer Inst. 27, 51–58 10.1016/j.jnci.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 183.Epple L.M., Griffiths S.G., Dechkovskaia A.M., Dusto N.L., White J., Ouellette R.J.et al. (2012) Medulloblastoma exosome proteomics yield functional roles for extracellular vesicles. PLoS ONE 7, e42064 10.1371/journal.pone.0042064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Osaki M. and Okada F. (2019) Exosomes and their role in cancer progression. Yonago Acta Med. 62, 182–190 10.33160/yam.2019.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Qin D., Yang W., Pan Z., Zhang Y., Li X. and Lakshmanan S. (2020) Differential proteomics analysis of serum exosomein burn patients. Saudi J. Biol. Sci. 27, 2215–2220 10.1016/j.sjbs.2020.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lv L., Li C., Zhang X., Ding N., Cao T., Jia X.et al. (2017) RNA profiling analysis of the serum exosomes derived from patients with active and latent Mycobacterium tuberculosis infection. Front. Microbiol. 8, 1051 10.3389/fmicb.2017.01051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Smith C.R., Batruch I., Bauça J.M., Kosanam H., Ridley J., Bernardini M.Q.et al. (2014) Deciphering the peptidome of urine from ovarian cancer patients and healthy controls. Clin. Proteomics 11, 23 10.1186/1559-0275-11-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Van Keer S., Pattyn J., Tjalma W.A.A., Van Ostade X., Ieven M., Van Damme P.et al. (2017) First-void urine: a potential biomarker source for triage of high-risk human papillomavirus infected women. Eur. J. Obstet. Gynecol. Reprod. Biol. 216, 1–11 10.1016/j.ejogrb.2017.06.036 [DOI] [PubMed] [Google Scholar]
- 189.Catani M.V., Savini I., Tullio V. and Gasperi V. (2020) The “Janus Face” of platelets in cancer. Int. J. Mol. Sci. 21, 788 10.3390/ijms21030788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jurasz P., Alonso-Escolano D. and Radomski M.W. (2004) Platelet–cancer interactions: mechanisms and pharmacology of tumour cell-induced platelet aggregation. Br. J. Pharmacol. 143, 819–826 10.1038/sj.bjp.0706013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lin R.J., Afshar-Kharghan V. and Schafer A.I. (2014) Paraneoplastic thrombocytosis: the secrets of tumor self-promotion. Blood 124, 184–187 10.1182/blood-2014-03-562538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Winkler J., Abisoye-Ogunniyan A., Metcalf K.J. and Werb Z. (2020) Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 11, 5120 10.1038/s41467-020-18794-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Clere N., Renault S. and Corre I. (2020) Endothelial-to-mesenchymal transition in cancer. Front. Cell Dev. Biol. 8, 10.3389/fcell.2020.00747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Snipsøyr M.G., Wiggers H., Ludvigsen M., Stensballe A., Vorum H., Poulsen S.H.et al. (2020) Towards identification of novel putative biomarkers for infective endocarditis by serum proteomic analysis. Int. J. Infect. Dis. 96, 73–81 10.1016/j.ijid.2020.02.026 [DOI] [PubMed] [Google Scholar]
- 195.Amati F., Vancheri C., Latini A., Colona V.L., Grelli S., D'Apice M.R.et al. (2020) Expression profiles of the SARS-CoV-2 host invasion genes in nasopharyngeal and oropharyngeal swabs of COVID-19 patients. Heliyon 6, e05143 10.1016/j.heliyon.2020.e05143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Blaabjerg M., Hemdrup A.L., Drici L., Ruprecht K., Garred P., Höftberger R.et al. (2018) Omics-based approach reveals complement-mediated inflammation in chronic lymphocytic inflammation with pontine perivascular enhancement responsive to steroids (CLIPPERS). Front. Immunol. 9, 741 10.3389/fimmu.2018.00741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang J., Wang N. and Xu A. (2018) Screening of genes associated with inflammatory responses in the endolymphatic sac reveals underlying mechanisms for autoimmune inner ear diseases. Exp. Ther. Med. 16, 2460–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Basu R.K., Standage S.W., Cvijanovich N.Z., Allen G.L., Thomas N.J., Freishtat R.J.et al. (2011) Identification of candidate serum biomarkers for severe septic shock-associated kidney injury via microarray. Crit. Care 15, r273 10.1186/cc10554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Posner M.G., Upadhyay A., Abubaker A.A., Fortunato T.M., Vara D., Canobbio I.et al. (2016) Extracellular fibrinogen-binding protein (Efb) from Staphylococcus aureus inhibits the formation of platelet-leukocyte complexes. J. Biol. Chem. 291, 2764–2776 10.1074/jbc.M115.678359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Satoh K., Hirayama T., Takano K., Suzuki-Inoue K., Sato T., Ohta M.et al. (2013) VacA, the vacuolating cytotoxin of Helicobacter pylori, binds to multimerin 1 on human platelets. Thrombosis J. 11, 23 10.1186/1477-9560-11-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mizrachi Nebenzahl Y., Blau K., Kushnir T., Shagan M., Portnoi M., Cohen A.et al. (2016) Streptococcus pneumoniae cell-wall-localized phosphoenolpyruvate protein phosphotransferase can function as an adhesin: identification of its host target molecules and evaluation of its potential as a vaccine. PLoS ONE 11, e0150320 10.1371/journal.pone.0150320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.van Eck N.J. and Waltman L. (2010) Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 84, 523–538 10.1007/s11192-009-0146-3 [DOI] [PMC free article] [PubMed] [Google Scholar]