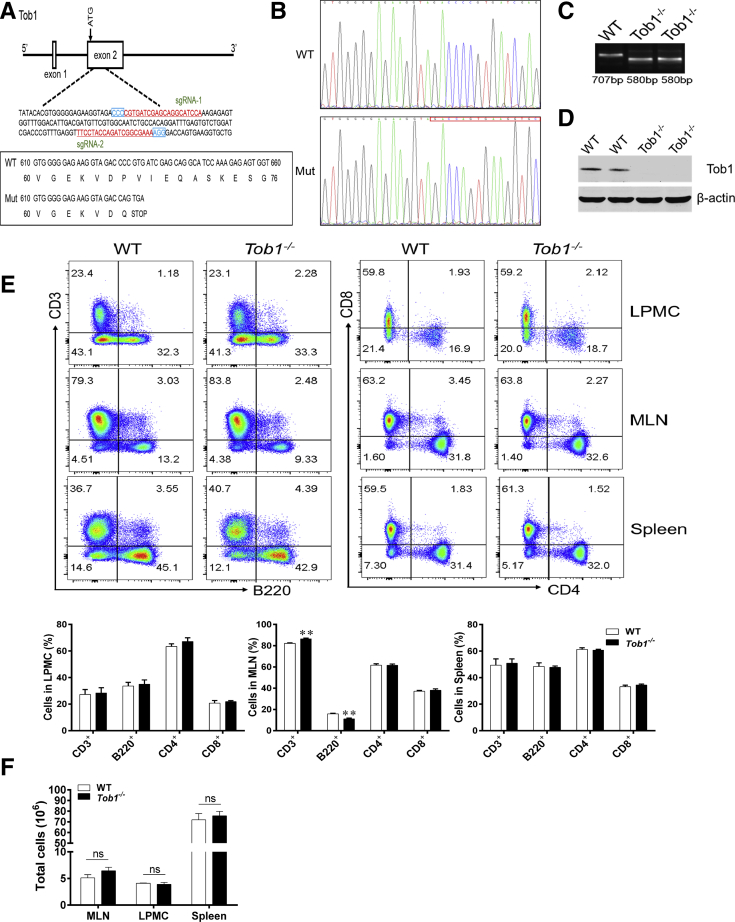
Figure 3.
The generation and genotyping of Tob1–/–mice. (A) Tob1–/– mice were generated using the CRISPR/Cas9 technique. The 2 single guide RNAs targeting the WT and Tob1 sequence are underlined and highlighted in red, and the protospacer adjacent motif sequences are labeled in blue. The reading frames of WT and mutant Tob1 sequence are listed below. The 127-bp deletion in Tob1 sequence results in an early stop codon TGA. (B and C) Target loci of Tob1 were amplified using genomic DNA templates from mouse tails of WT and Tob1–/– mice. PCR products of the targeted fragment were subjected to (B) sequencing and (C) agarose gel electrophoresis, indicating the 127 missing bp in the Tob1–/– mice. (D) Protein levels of Tob1 in the colon tissues of WT littermates and Tob1–/– mice were detected by Western blotting, and β-actin was used as endogenous reference gene. (E) LPMCs and single-cell suspension of MLNs and spleens were obtained from 6- to 8-week-old WT littermates (n = 4) and Tob1–/– mice (n = 4), respectively, and then stained with fluorochrome-conjugated anti-CD3, anti-CD4, anti-CD8, and anti-B220 mAbs, respectively. The frequencies of B220+ B cells, CD3+ T cells (gated on total cells), and CD4+ and CD8+ T cells (gated on CD3+ T cells) in LPMCs, MLNs, and spleens, respectively, were analyzed by flow cytometry. Percentages of these cells in LPMCs, MLNs, and spleens were shown in the bar chart, respectively. (F) The total cells in MLN, LPMC, and spleen were calculated and are presented in the bar chart. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗∗P < .01 vs WT control animals from the same group.