Abstract
Based on predictable, complementary base pairing, DNA can be artificially pre-designed into versatile DNA nanostructures of well-defined shapes and sizes. With excellent addressability and biocompatibility, DNA nanostructures have been widely employed in biomedical research, such as bio-sensing, bio-imaging, and drug delivery. With the development of the chemical biology of nucleic acid, chemically modified nucleic acids are also gradually developed to construct multifunctional DNA nanostructures. In this review, we summarize the recent progress in the construction and functionalization of chemically modified DNA nanostructures. Their applications in the delivery of chemotherapeutic drugs and nucleic acid drugs are highlighted. Furthermore, the remaining challenges and future prospects in drug delivery by chemically modified DNA nanostructures are discussed.
Keywords: self-assembly, nucleic acid nanostructure, chemical modification, drug delivery, tumor therapy
Graphical abstract
Public summary
-
•
With excellent addressability and biocompatibility, DNA nanostructures are promising candidates for bio-sensing, bio-imaging, and drug delivery
-
•
The recent progress in chemical modifications of DNA nanostructures is summarized
-
•
Chemically modified DNA nanostructures for efficient delivery of chemotherapeutic drugs and nucleic acid drugs are highlighted
-
•
Challenges and prospects of future development toward chemically modified DNA nanostructures for drug delivery are discussed
Introduction
Most known as a carrier of genetic information, DNA can also be employed as a building block for constructing versatile DNA nanostructures by complementary base pairing. Different self-assembly strategies can create DNA nanostructures with controllable sizes and shapes. Meanwhile, various stimuli-responsive components can be site-specifically organized in the versatile DNA nanostructures through non-covalent or covalent binding. Due to its excellent structural programmability, spatial addressability, and biocompatibility, DNA self-assembled nanostructures have been widely applied in numerous research fields, such as information storage, smart nanodevices, biosensors, and drug delivery.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 The wide-ranging DNA nanostructures have developed into one of the most representative self-assembly systems with a long and noteworthy history.
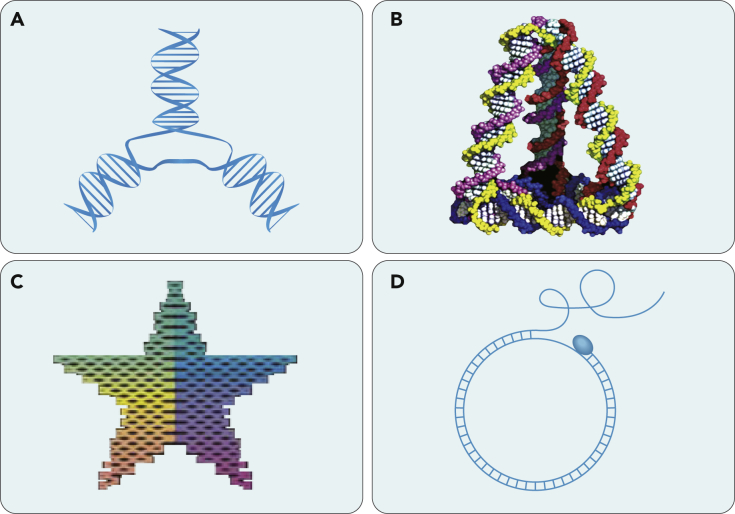
In the early 1980s, Nadrian C. Seeman, the pioneer of DNA nanotechnology, proposed that nucleic acids could be utilized as building blocks for the assembly of nanoscale architectures by Watson-Crick base pairing (Figure 1A).15 Subsequently, a series of two-dimensional (2D) DNA nanostructures were successfully constructed.16, 17, 18, 19, 20, 21, 22, 23 Meanwhile, DNA polyhedral nanostructures with 3D morphologies were efficiently assembled in high yields (Figure 1B).24, 25, 26, 27 In 2006, Rothemund developed a strategy to fold a long, single-stranded circular scaffold DNA, with hundreds of short staple strands, into various 2D nanostructures, termed “DNA origami” (Figure 1C).28 Based on this technology, a number of interesting DNA origamis have been rationally designed and fabricated.29, 30, 31, 32, 33, 34, 35, 36, 37 In addition, rolling circle amplification (RCA) has been employed to construct various DNA hydrogels (Figure 1D).38 These structurally well-defined DNA nanostructures have been further developed to spatially arrange a variety of functional components with high precision.39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49 Besides ordinary, natural nucleic acids, chemically modified DNA also has played an essential role in the rational structural assembly.
Figure 1.
Representative strategies for the assembly of DNA nanostructures
(A) DNA junctions. Reproduced with permission from Seeman15 Copyright 1982, Elsevier.
(B) DNA tetrahedron. Reproduced with permission from Goodman et al.24 Copyright 2005, American Association for the Advancement of Science.
(C) DNA origami. Reproduced with permission from Rothemund28 Copyright 2006, Nature Publishing Group.
(D) DNA nanostructures based on rolling circle amplification. Reproduced with permission from Ali et al.38 Copyright 2014, Royal Society of Chemistry.
Chemical modification of DNA
Various organic reactions have been developed for the chemical modification of DNA.50,51 Based on additional covalent bonds, the chemically modified DNA can facilitate structural assembly and cargo loading. Meanwhile, chemical modification can also protect DNA nanostructures from enzymatic digestion to enhance stability. Due to these advantages, chemically modified DNA has attracted much attention in biosensors, bio-imaging, and drug delivery.
To expand the scope of DNA nanostructure applications, different modification sites have been studied, such as the two terminal, phosphate skeleton, bases, and sugar ring (Figure 2). Terminal-modified DNA (containing amino, thiol, azide, or alkyne groups) can conjugate with branched molecules to construct branched DNA for the subsequent assembly of crosslinked materials.52 The phosphate skeleton can be modified by thiophosphate to enhance its stability and introduce additional reactive sites for further functionalization.53,54 Bases are often modified by floxuridine, a typical chemotherapeutic drug.55 The representative modification of the sugar ring is mirror DNA (L-DNA), which can significantly improve the molecule's enzymatic stability to facilitate drug delivery.56,57 All of these above-mentioned modified nucleotides are commercially available. These modified DNA strands can be easily and efficiently obtained by solid-phase synthesis. Meanwhile, the modified nucleotides can also be incorporated into DNA by DNA polymerase enzymes.
Figure 2.
Chemical modifications of DNA for assembly of DNA nanostructures
(1) 5′ and 3′ terminal (red) modifications (such as amino, thiol, azide, and alkyne); (2) phosphate skeleton (purple) modifications (such as PS); (3) base (green) modifications (such as floxuridine); (4) sugar ring (blue) modifications (such as L-DNA).
Since chemically modified DNAs are all made commercially available by solid-phase synthesis, the technique routes and product yields are good enough for broad applications in biomedical research. These obtained chemically modified DNAs still retain the ability of strict complementary base pairing to construct versatile DNA nanostructures. These above-mentioned different chemical modification strategies can be flexibly utilized to solve specific problems in different situations. For example, the terminal-modified DNA with high conjugation efficiency is often employed in the fabrication of branched DNA structures to assemble hydrogels or nanoparticles. The DNA modified with thiophosphate at arbitrary sites of the phosphate skeleton is often adopted to enhance stability. Floxuridine-incorporated nucleotides are usually directly employed as building blocks of wireframe DNA nanostructures or monomers for RCA to prepare drug-loaded nanoflowers. With its modified sugar ring, the mirror DNA is always directly employed for the co-assembly of versatile DNA nanostructures with much-enhanced stability. These diverse chemical modification strategies are vital to the development of DNA nanotechnology for biomedical applications.
Chemically modified DNA nanostructures
Chemically modified DNAs have been widely employed as the building blocks for the construction of versatile DNA nanostructures. Various strategies, such as terminal covalent modifications, the branched PCR and ligation, and direct covalent modifications onto DNA origami, have been developed to assemble chemically modified DNA nanostructures.
Terminal covalent modifications
The covalent coupling reaction between the terminal-modified DNA and branched organic molecules with different numbers of functional groups is a typically adopted strategy for the construction of branched DNA nanostructures with indicated numbers of DNA arms. These branched DNA nanostructures can function well as the building blocks for the co-assembly of higher-order DNA nanostructures with pre-designed sizes and shapes.
In 1999, Scheffler et al. synthesized one pair of trisoligonucleotidyls, with cDNA sequences, through solid-phase synthesis based on the flexible branched organic molecule (4-amino-4-(3-hydroxypropyl)heptane-1,7-diol, Figure 3A).58 Each double-stranded DNA between the two complementary trisoligonucleotidyls was described as a chemical C-C bond. The two main possible topologies for the DNA assemblies were denoted as nano-acetylene (assembly between one pair of trisoligonucleotidyls) and nano-cyclobutadiene (assembly between two pairs of trisoligonucleotidyls). The co-assembly models proposed in this work inspired subsequent research based on covalently conjugated branched DNA. In 2008, Stepp et al. constructed a pair of small-molecule-DNA hybrids with a rigid tris(phenylacetylene) core to organize three DNA strands at 120° intervals.59 They found that the complementary hybrids can form a caged dimer (nano-acetylene, as mentioned above) with a higher Tm in dilute solution and the DNA aggregate with a lower Tm in concentrated solution. In 2012, Eryazici et al. employed a synthetic route similar to the one reported by Stepp et al. (both 3′ modifications for conjugation) to construct another pair of complementary hybrids in reverse directions (including 3′ and 5′ modifications).60 This pair of branched DNA can form a face-to-face dimer at a wide concentration range of the DNA arms. This work also reveals that hydrophobic interaction among the rigid cores of organic molecules plays an essential role in assembling the chemically modified DNA nanostructures. In 2015, Hong et al. replaced the rigid aromatic organic cores described in the report of Stepp et al. with flexible, small-molecule cores.61 These well-defined cage dimers can be assembled at DNA concentrations as high as 102 μM with oligonucleotides of moderate length (<20 bp). This report further confirms that the organic molecular core is crucial for the efficient formation of chemically modified DNA cage dimers. Meanwhile, in a subsequent study, Hong et al. constructed a pair of complementary tetrahedral small-molecule-DNA hybrids with a rigid organic core to assemble DNA-based polymeric nanoparticles.62 The sizes and distributions of these DNA nanoparticles were easily modulated by changing the concentration of branched DNA, the assembly time, and the salt concentration. An enhanced cellular uptake was observed in this DNA-based nanoparticle, demonstrating the potential application of this strategy in drug delivery.
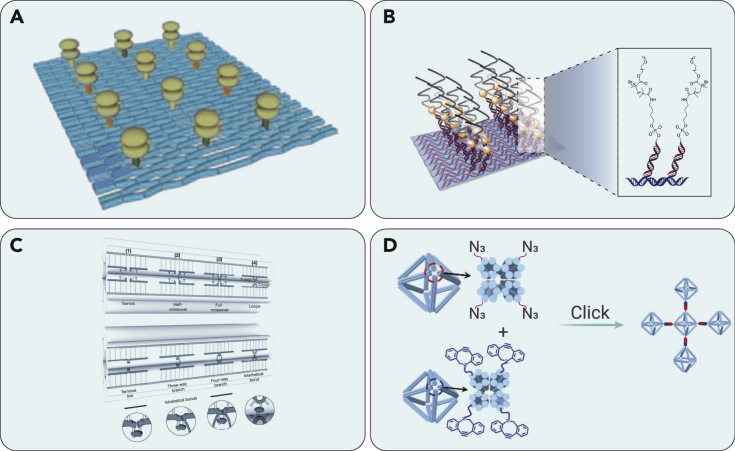
Figure 3.
Representative DNA nanostructures assembled by terminal covalently modified DNA
(A) Self-assembly of trisoligonucleotides. Reproduced with permission from Scheffler et al.58 Copyright 1999, Wiley-VCH.
(B) Self-assembly of a DNA hexagon as a template for the organization of gold nanoparticles. Reproduced with permission from Aldaye et al.63 Copyright 2006, Wiley-VCH.
(C) Guest-mediated access to a single DNA nanostructure. Reproduced with permission from Aldaye et al.64 Copyright 2007, American Chemical Society.
(D) Structurally switchable 3D DNA assemblies. Reproduced with permission from Aldaye et al.65 Copyright 2007, American Chemical Society.
(E) Self-assembly of a DNA dodecahedron from 20 trisoligonucleotides. Reproduced with permission from Zimmermann et al.66 Copyright 2008, Wiley-VCH.
(F) Metal-nucleic acid cages. Reproduced with permission from Yang et al.67 Copyright 2009, Nature Publishing Group.
(G) DNA nanotubes with a longitudinal variation. Reproduced with permission from Lo et al.68 Copyright 2010, Nature Publishing Group.
(H) DNA-imprinted polymer nanoparticles. Reproduced with permission from Trinh et al.69 Copyright 2018, Nature Publishing Group.
In 2006, Aldaye et al. prepared six DNA oligonucleotides with rigid organic molecules as a central linker through solid-phase synthesis (Figure 3B).63 These six DNA oligonucleotides can hybridize with each other hand by hand, to construct a well-defined cyclic hexamer. Due to the addressability of the DNA hexamer, six gold nanoparticles can be efficiently arranged at the pre-designed terminals of the DNA sequences to form a crown-like structure. Various multifunctional nanoparticles can be spatially positioned in this precisely organized pattern. In the following year, Aldaye et al. fabricated a pair of DNA dimers with complementary sequences for co-assembly (Figure 3C).64 In this system, an additional small molecule (Ru(bpy)32+) was included as an auxiliary compound (guest) to facilitate assembly of a single DNA nanostructure (DNA square) and avoid the possible random assembly of a library of products. Similarly, a pair of DNA trimers was prepared to construct ladder-like structures with the addition of (Ru(bpy)32+), which were further assembled into large DNA fibers of over tens of micrometers. This strategy, based on the simple use of a small molecule to reprogram DNA assemblies, is a promising approach to control DNA self-assembly to achieve desired sizes and shapes.
For the construction of 3D DNA frameworks, in 2007, Aldaye et al. synthesized a series of cyclic single-stranded DNA building blocks (including triangle, square, pentagon, and hexamer) through solid-phase synthesis with 3′ phosphate modification and subsequent chemical ligation (Figure 3D).65 These cyclic DNA structures can function as scaffolds to co-assemble with helper strands to construct structurally switchable 3D DNA assemblies. This work attracted much attention, leading to the efficient construction of versatile 3D DNA nanostructures in subsequent studies. In 2008, Zimmermann et al. further prepared 20 trisoligonucleotidyls, with different pre-designed DNA sequences, through solid-phase synthesis using C3h linkers (Figure 3E).66 These trisoligonucleotidyls can hybridize with each other in a pre-designed manner to construct a 3D DNA dodecahedron, demonstrating a promising option for constructing 3D DNA framework nanostructures with arbitrary morphologies.
In 2009, Yang et al. introduced some organic chelating sites into a DNA prism through solid-phase synthesis to precisely organize the metal ions (Figure 3F).67 Various metal ions, such as Cu+, Ag+, Zn2+, Co2+, Cd2+, Au+, and Eu2+, can be precisely incorporated into the indicated chelating sites to form a metal-DNA 3D cage. This metal-DNA hybrid nanostructure exhibited a much-enhanced resistance against chemical and thermal denaturation. This work promoted the development of metal-DNA hybrid nanostructures for applications in the fields of bio-sensing. In 2010, Lo et al. fabricated a series of DNA nanotubes, with longitudinal variation and capsules along the tube length, through co-assembly of covalently conjugated cyclic DNA and helper strands (Figure 3G).68 These DNA nanotubes can precisely position gold nanoparticles of an appropriate size in the separate capsules during the co-assembly process. The loaded gold nanoparticles can be efficiently released after adding a competitive DNA strand with fully complementary sequences. This controllable and stimuli-responsible loading and release strategy based on DNA nanostructures may be further applied in drug delivery systems. Based on the addressability of DNA nanostructures, in 2018, Trinh et al. developed a method to transfer positional information from a DNA cube to a polymeric nanoparticle (Figure 3H).69 The polymeric nanoparticle encapsulated inside the cube was fabricated through a covalent crosslinking reaction. After denaturing, the particles imprinted with different DNA strands can be efficiently constructed. This work provided a general strategy to obtain non-centrosymmetric nanostructures predictably.
Branched PCR and ligation
PCR is a powerful molecular biological tool to obtain long, targeted, double-stranded DNA on a large scale. In theory, versatile DNA nanostructures with multiple DNA arms can be constructed by quickly altering traditional linear DNA primer pairs into covalently conjugated branched DNA (denoted “branched PCR”). Based on the branched PCR system, various functional components can be efficiently incorporated into long, double-stranded DNA networks by arbitrary chemical modification of the branched primer pair.
In 2008, Lee et al. synthesized a dimer of a forward primer through an amide-coupling reaction between a 5′ amide-modified single-stranded DNA primer and an organic molecular linker (Figure 4A).70 Based on traditional PCR, the dimer of the forward primer and a normal linear reverse primer were combined to amplify long, double-stranded DNA. The triblock DNA nanostructures (long dsDNA-organic molecule-long dsDNA) were clearly visible by atomic force microscope (AFM) imaging. Meanwhile, Keller et al. prepared trimers of primers in the same year through solid-phase synthesis (Figure 4B).71 One pair of trimers was directly introduced into the branched PCR system as a primer pair. After branched PCR, the DNA network was clearly apparent by AFM imaging. These DNA network meshes can be easily tunable by changing the length of the DNA template. This work provides an alternative strategy for the efficient construction of double-stranded DNA networks. Furthermore, in 2011, Lee et al. prepared the dimers and trimers of DNA oligonucleotides by three cross-coupling reactions, concluding that the amide-coupling reaction can achieve the highest yield (Figure 4C).72 The obtained branched DNA can be elongated through hybridization with the overhangs of long double-stranded DNA and a subsequent ligation process. This work provided a method for the construction of micrometer-sized DNA nanostructures. In 2016, Liu et al. synthesized a trimer of DNA oligonucleotides through the thiol-Michael addition reaction.73 This trimer can hybridize with single-stranded DNA to form a DNA junction with four base overhangs (GGGG). The covalently conjugated DNA junction can efficiently assemble with an EGFP gene containing single-stranded overhangs (CCCC, obtained by digestion with the restriction endonuclease, BbsI). After ligation, a stable gene nanoparticle was successfully constructed for eukaryotic expression. This report demonstrates a promising strategy based on using a chemically modified branched DNA nanostructure as a crosslinker to construct a nanoparticle system for gene delivery efficiently.
Figure 4.
Representative DNA nanostructures constructed by branched PCR and ligation
(A) Synthesis of DNA-organic molecule-DNA triblock oligomers as the primer for PCR. Reproduced with permission from Lee et al.70 Copyright 2008, American Chemical Society.
(B) DNA polymerase-catalyzed DNA network growth. Reproduced with permission from Keller et al.71 Copyright 2008, American Chemical Society.
(C) Elongation of branched oligomers by ligation. Reproduced with permission from Lee et al.72 Copyright 2011, American Chemical Society.
(D) Thermostable branched DNA nanostructures as the primers for PCR. Reproduced with permission from Hartman et al.74 Copyright 2013, Wiley-VCH.
Besides the direct crosslinking of terminal-modified single-stranded DNA, other organic reactions have also been employed to bundle up the double-stranded DNA. In 2013, Hartman et al. employed a light-induced crosslinking reaction between a thymine base (T) and the psoralen molecule to form a covalently crosslinked T psoralen-T unit (Figure 4D).74 After crosslinking, thermostable branched double-stranded DNA with single-stranded overhangs was efficiently synthesized. This type of branched DNA structure can also be easily adopted as the modular primer pair in a branched PCR system. After including a GFP gene as the template, an extended DNA network can be efficiently fabricated for subsequent gene expression.
Direct covalent modifications onto DNA origami
DNA origami is often constructed by co-assembly of a long single-stranded circular DNA scaffold (e.g., bacteriophage M13mp18, 7,249 nt) and multiple short DNA staple strands in a one-pot system. With the developments in chemical modification of DNA, various attempts have been carried out to directly modify the DNA origami in situ to construct multifunctional DNA nanostructures. A series of efficient chemical reactions have been developed for site-specific modification of DNA origami with high yield.
In 2010, Voigt et al. employed a rectangular-shaped DNA origami as a highly versatile breadboard for the investigation of surface chemical reactions with single-molecule resolution (Figure 5A).75 These chemical reactions, such as click chemistry and amide coupling, can be generally performed at pre-designed positions on the DNA origami template. Based on the specific interaction between biotin (attached to the organic reaction groups) and streptavidin, these reactions on the surface of DNA origami can be directly imaged by AFM. This work demonstrates that it is possible for site-specific chemical modification of DNA origami in a post-assembly manner. Furthermore, in 2016, Tokura et al. employed DNA origami as a template to site-specifically organize initiators conjugated on the terminals of DNA strands for in situ atom-transfer radical polymerization (Figure 5B).76 Based on the addressability of DNA origami, differently nanopatterned polymers of various heights can be efficiently obtained with high precision. After crosslinking, the polymeric nanostructures can remain stable in solution without the support of the DNA origami. In principle, the judicious utilization of this DNA origami strategy enables the fabrication of polymeric nanostructures with arbitrary patterns.
Figure 5.
Representative chemically modified DNA origami
(A) Single-molecule chemical cleavage and coupling reactions on DNA origami. Reproduced with permission from Voigt et al.75 Copyright 2010, Nature Publishing Group.
(B) Bottom-up fabrication of nanopatterned polymers on DNA origami by in situ atom-transfer radical polymerization. Reproduced with permission from Tokura et al.76 Copyright 2016, Wiley-VCH.
(C) Sequence-programmable covalent bonding of DNA origami in a site-selective and scalable manner. Reproduced with permission from Gerling et al.77 Copyright 2018, American Association for the Advancement of Science.
(D) Covalent-bound DNA origamis obtained by cupper-free click reaction. Reproduced with permission from Lin et al.78 Copyright 2019, American Chemical Society.
To enhance the stability of DNA origami, in 2018, Gerling et al. created an additional covalent cyclo-butane pyrimidine dimer between two adjacent thymidines via UV irradiation (Figure 5C).77 These reactive sites for covalent conjugation can be artificially pre-designed in the desired positions. After irradiation, the stabilized DNA origami remains intact at high temperatures and in highly purified water without additional cations. Notably, enhanced resistance against digestion by DNA nuclease was also observed in these chemically stabilized DNA origamis. This report provides a general strategy for stabilizing DNA origami to promote the broad application of DNA nanostructures in various research fields. In 2019, Lin et al. employed a click chemical reaction to manipulate a series of octahedral DNA origamis to efficiently construct higher-order DNA origami (Figure 5D).78 Azide and alkyne reaction groups were site-specifically anchored onto desired vertices of octahedral DNA origami. After click reaction, various super-DNA origamis with different architectures were obtained by changing reactive valence or stoichiometric ratios of the chemically modified octahedral DNA origami. This work creatively treats the entire octahedral DNA origami as a molecular backbone for a direct chemical reaction, mimicking traditional organic molecular synthesis, making it possible to construct various super-DNA nanostructures with complex architectures.
Besides these direct covalent modifications onto DNA origami, some chemically modified DNA origamis have also been constructed by assembling pre-modified staple strands or hybridizing covalently modified DNA strands. In 2013, Burns et al. constructed a self-assembled small tubular DNA origami to function as a nanopore in lipid bilayers.79 The chemical modification of the phosphate backbone with hydrophobic alkyl groups in the pre-selected middle region of the tubular DNA origami can mask the negative charge present in unmodified DNA. With an outer hydrophobic belt, the DNA nanopores could efficiently span the lipid bilayer to support a constant transmembrane current. This work proves that rationally designed, bottom-up DNA nanostructures can be successfully employed as macromolecules that mimic the biological functions of naturally existing proteins. In 2015, Knudsen et al. synthesized a series of organic polymer wires containing multiple short DNA handles through in situ solid-phase synthesis.80 The DNA-modified soft and bendable polymer can be artificially organized onto the surface of two- and 3D DNA origamis in a pre-designed route through DNA hybridization. Meanwhile, the precisely positioned organic polymers possess a π-conjugated backbone, displaying a range of electronic properties. This report provides a promising strategy to organize individual functional polymer in pre-designed patterns to construct molecular-scale electronic nanodevices.
Drug delivery
Various chemotherapeutic drugs and nucleic acid drugs have been developed to combat different diseases. Systemic delivery of these drugs is challenging because of these existing limiting factors, such as solubility, stability, targeting transportation, membrane penetration, and controllable release. Numerous versatile nanocarriers, including liposomes, polymers, and inorganic nanoparticles, have been constructed. Considering the possible cytotoxicity and immunogenicity of these carriers, the wide application of these carriers is limited. Fortunately, various chemical modifications of nucleic acids can be employed to construct versatile chemically modified DNA nanostructures. Drug delivery is a promising application of these chemically modified DNA nanostructures with their enhanced stability and attached functional moieties by chemical modifications. Indeed, several rationally designed drug delivery systems have been reported on the basis of chemically modified DNA nanostructures for the systemic delivery of chemotherapeutic drugs and nucleic acid drugs.
Delivery of chemotherapeutic drugs
Chemically modified wireframe DNA nanocarriers
Chemically modified wireframe DNA nanostructures with precise control of sizes and shapes have been successfully developed to deliver chemotherapeutic drugs through base intercalation or site-specific chemical conjugation. These uniform DNA nanostructures can organize drug molecules in a site-specific manner to achieve a structurally well-defined drug delivery system. Efficient cellular uptake can be achieved by tuning the types of DNA nanostructures and attached targeting elements. Meanwhile, controllable drug release behavior can be easily realized on the basis of the chemically modified wireframe DNA nanostructures by incorporating stimuli-responsive chemical bonds or nucleic acid motifs. These modifications involve functional components that can be stimuli-responsively cleaved or opened in specific environments (including reduction, oxidation, and acidification) and/or by targeting molecular triggers (such as proteins and nucleic acids) for efficient controllable release.
In 2016, Kim et al. constructed a mirror DNA tetrahedron by co-assembly of four pre-designed, single-stranded L-DNA for the loading of the chemotherapeutic drug, doxorubicin (DOX); (Figure 6A).81 The mirror DNA tetrahedron exhibited greater serum stability than the normal D-DNA tetrahedron. A prolonged dwell time of the mirror DNA tetrahedron in the tumor region played an important role in the enhanced therapeutic effects in vivo. This work demonstrates a significant application of chemically modified mirror DNA nanostructures in the field of drug delivery systems. Furthermore, in 2019, the same group constructed a library of mirror DNA nanostructures with different sizes and shapes to load DOX for drug delivery.82 They found that rational design of the size and shape of chemically modified DNA nanostructures are essential for successful cellular uptake and macrophage evasion. Finally, a pyramid mirror DNA cage structure demonstrated the highest tumor specificity and resulted in an efficient therapeutic effect.
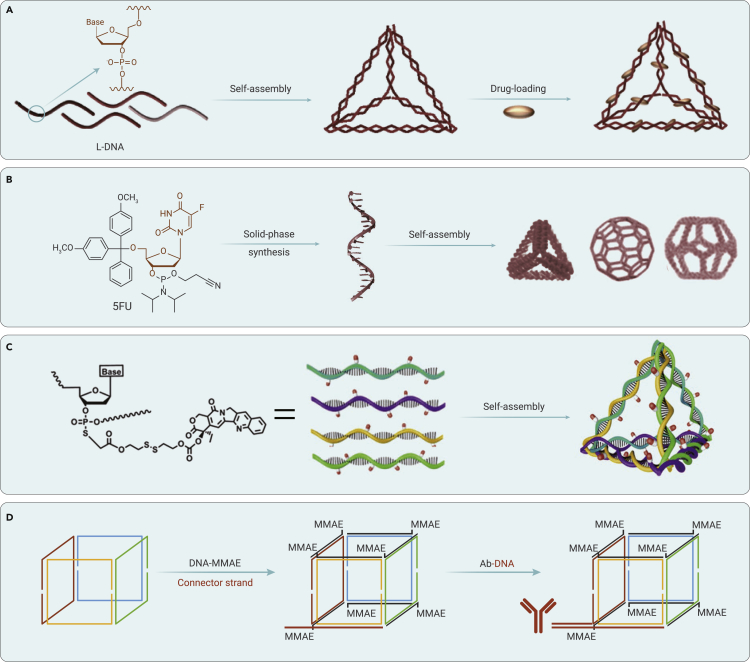
Figure 6.
Delivery of chemotherapeutic drugs based on chemically modified wireframe DNA nanostructures
(A) Mirror DNA tetrahedron for tumor-specific delivery of DOX. Reproduced with permission from Kim et al.81 Copyright 2016, Elsevier.
(B) Floxuridine-containing DNA polyhedra for cancer therapy. Reproduced with permission from Mou et al.83 Copyright 2017, Wiley-VCH.
(C) Camptothecin-grafted DNA tetrahedron for cancer therapy. Reproduced with permission from Zhang et al.84 Copyright 2019, Wiley-VCH.
(D) Antibody-conjugated Wireframe DNA Cube for targeted delivery of MMAE. Reproduced with permission from Märcher et al.85 Copyright 2021, Wiley-VCH.
To precisely load chemotherapeutic drugs in DNA nanostructures, in 2017, Mou et al. fabricated a library of well-defined DNA polyhedra with the incorporation of floxuridine-integrated single-stranded DNA by solid-phase synthesis (Figure 6B).83 The drug loading ratios can be facilely controlled by changing the DNA sequences. They also found that the morphology of these DNA polyhedra can affect their cellular uptake. The floxuridine-loaded buckyballs exhibited effective tumor growth inhibition in vivo. On the basis of this strategy, various other nucleoside analogs can be included in the DNA strands for the co-assembly of structurally well-defined DNA nanodrugs. Furthermore, in 2019, Zhang et al. employed carbon ethyl bromide-modified camptothecin (CPT) to conjugate with phosphorothioate-modified DNA strands for the self-assembly of chemically modified DNA tetrahedrons (Figure 6C).84 The loading ratio and site of CPT molecules in the DNA nanostructure can be easily tuned. The obtained CPT-tetrahedron nanostructure exhibited efficient cellular uptake and marked inhibition of tumor growth in a tumor-bearing mouse model. Moreover, in 2020, Zhang et al. also employed CPT-conjugated DNA oligonucleotides to form Y-shaped DNA motifs for the construction of an injectable CPT-conjugated DNA hydrogel for local chemotherapy.86 This CPT-loaded DNA hydrogel demonstrated high therapeutic efficacy against tumor recurrence based on the long-term cytotoxicity of the residual tumor cells. These reports provide a general strategy for site-specific conjugation of chemotherapeutic drugs on uniformed DNA nanostructures.
For target delivery of a drug-conjugated wireframe DNA nanostructure, in 2021, Märcher et al. organized an antibody onto a chemotherapeutic drug monomethyl auristatin E (MMAE) (a tubulin inhibitor)-loaded wireframe DNA cube (Figure 6D).85 The MMAEs were covalently conjugated to single-stranded DNA via a cleavable organic linker. The single-stranded DNA-conjugated antibody was efficiently attached to the remaining vertex of the seven MMAE-loaded DNA cube through efficient DNA hybridization. In this case, the drug-to-antibody ratio is tuned precisely to 7:1. Such precision is typically difficult to realize in traditional antibody-drug conjugates. The constructed antibody (trastuzumab)-DNA cube-MMAE exhibited an efficient target in HER2-positive SKBR3 cells in vitro. This report demonstrates a representative example of employing uniformed DNA nanostructures as a template for the site-specific loading of chemotherapeutic drugs and targeting groups.
Chemically modified DNA spherical nanocarriers
Most chemotherapeutic drugs are hydrophobic. Before administration, the efficient conversion of these hydrophobic molecules into a hydrophilic form is essential. The DNA strand possesses excellent hydrophilicity and unique programmability. After conjugation between hydrophobic drug and hydrophilic DNA, chemically modified drug-DNA hybrid nanoparticles can be successfully constructed with a 3D-structured core (hydrophobic drug) and shell (hydrophilic DNA). Such hybrid spherical nanoparticles are efficiently internalized by tumor cells for drug delivery.
In 2018, Yang et al. synthesized a DNA aptamer-grafted, photo-responsive hyperbranched polymer using click reaction (Figure 7A).87 The hydrophilicity of the hyperbranched polymer was blocked by a nitrobenzyl group to result in a hydrophobic core. The hydrophobic chemotherapeutic drug, DOX, was then successfully loaded into the hydrophobic core. This aptamer-coated nanoparticle can be efficiently internalized by target cells. After UV irradiation, the nitrobenzyl group was cleaved off from the hyperbranched polymer to expose the hydrophilic carboxylic acid group for controllable disassembly. The loaded DOX was gradually released into the cytoplasm during the disassembly process for targeted and stimuli-responsive chemotherapy. This work demonstrates that the chemical conjugation of hydrophobic molecules onto the terminal of the hydrophilic DNA strand can form a hybrid nanoparticle as a carrier for drug delivery.
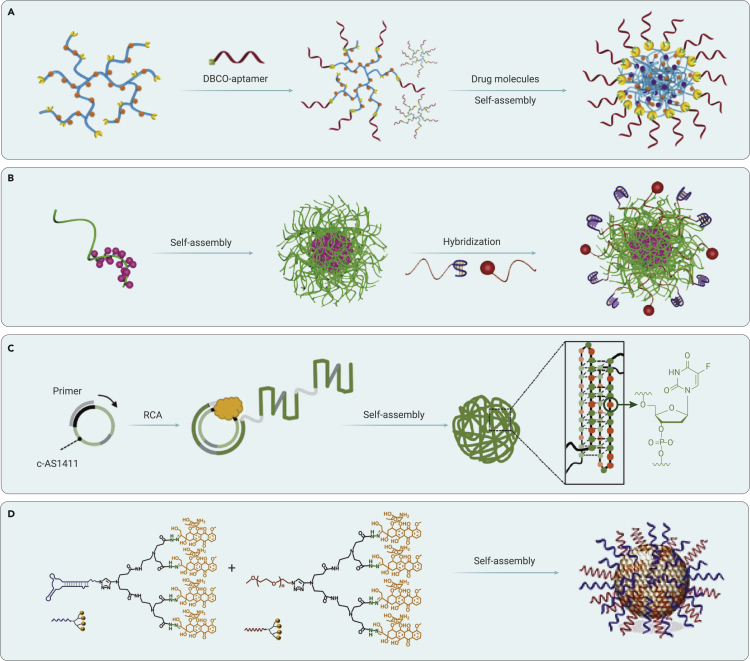
Figure 7.
Delivery of chemotherapeutic drugs based on chemically modified DNA spherical nanoparticles
(A) Aptamer-grafted hyperbranched polymer nanocarrier for targeted and photo-responsive drug delivery of DOX. Reproduced with permission from Yang et al.87 Copyright 2018, Wiley-VCH.
(B) Grafting PTX onto phosphorothioate oligonucleotides for drug delivery. Reproduced with permission from Guo et al.88 Copyright 2019, Wiley-VCH.
(C) Self-assembly of polymerized floxuridine-incorporated aptamer through RCA. Reproduced with permission from Tran et al.89 Copyright 2020, Royal Society of Chemistry.
(D) Multivalent aptamer-drug conjugates for drug delivery. Reproduced with permission from Geng et al.90 Copyright 2021, Wiley-VCH.
To precisely load chemotherapeutic drugs, in 2019, Guo et al. site-specifically grafted benzyl bromide-modified paclitaxel (PTX) onto a phosphorothioate-modified DNA backbone (Figure 7B).88 The amphiphilic PTX-DNA hybrid structure can efficiently assemble into a spherical nucleic acid-like micellar nanoparticle. A targeting aptamer and functional antisense strand can be readily integrated into this nanoparticle by hybridization and sequence design. This multifunctional nanoparticle demonstrates active targeting delivery and efficient inhibition of tumor growth in vivo. Furthermore, in 2020, Ren et al. replaced the PTX with cisplatin to synthesize an aptamer/poly T-Pt drug conjugate for chemotherapy.91 This hybrid DNA conjugate also self-assembles into a spherical nucleic acid-like nanoparticle to achieve targeted chemotherapy in vivo. These reports provide additional strategies to load and deliver hydrophobic chemotherapeutic drugs in a site-specific manner. Similarly, in 2020, Zhu et al. from the same group grafted two PTX molecules onto the terminal of a floxuridine-integrated antisense DNA strand.92 This PTX-antisense hybrid structure can also form spherical nucleic acid-like micellar nanoparticles. This multifunctional nanoparticle achieved synergistic tumor therapy by combining chemotherapy (PTX and floxuridine) and gene therapy (antisense targeting P-gp).
In 2020, Tran et al. integrated floxuridine into the AS1411 aptamer through RCA to form a drug-aptamer hybrid nanoparticle (Figure 7C).89 The floxuridine-incorporated AS1411 aptamer maintained its efficient cell-targeting ability. This nanoparticle demonstrated enhanced serum stability and marked tumor-targeted accumulation. After treatment, a synergistic therapeutic effect, depending on the upregulation of p53 by AS1411 and downregulation of thymidylate synthase by floxuridine, was observed. This report provides a general method, based on RCA, to introduce various nucleoside-incorporated, aptamer-drug conjugates for actively targeted drug delivery in vivo.
To combine chemotherapy and immunotherapy, in 2021, Geng et al. grafted four DOX molecules onto the terminal of a DNA aptamer by acid-labile linkers (Figure 7D).90 This amphiphilic DOX-aptamer hybrid can co-assemble into nanomicelles. After mixing the DOX-PEG hybrid at an optimized ratio with DOX-aptamer, a balance of blood circulation stability and tumor-targeting were achieved. The obtained nanomicelles could release DOX in the acidic tumor microenvironment selectively. After co-administration of α-PD1, an enhanced tumor-specific immunotherapy effect was observed in an immunocompetent, tumor-bearing mouse model. This report demonstrates that tumor-targeting, drug-DNA nanomicelles can be employed as a nanoplatform to improve the response rate of immunotherapy.
Delivery of nucleic acid drugs
Based on different gene therapy systems, such as antisense, RNA interference (RNAi), gene expression, and gene editing, various nucleic acid drugs have been developed for the treatment of diseases in various situations.93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105 However, the delivery of nucleic acid drugs to target organelles remains a challenge, limiting application in the clinic. Fortunately, due to inherent homology, nucleic acid drugs (including antisense, siRNA, shRNA, sgRNA, mRNA, and gene expression cassettes) can be facilely loaded by DNA nanocarriers through base pairing hybridization or chemical covalent crosslinking.106, 107, 108, 109, 110, 111, 112, 113, 114, 115
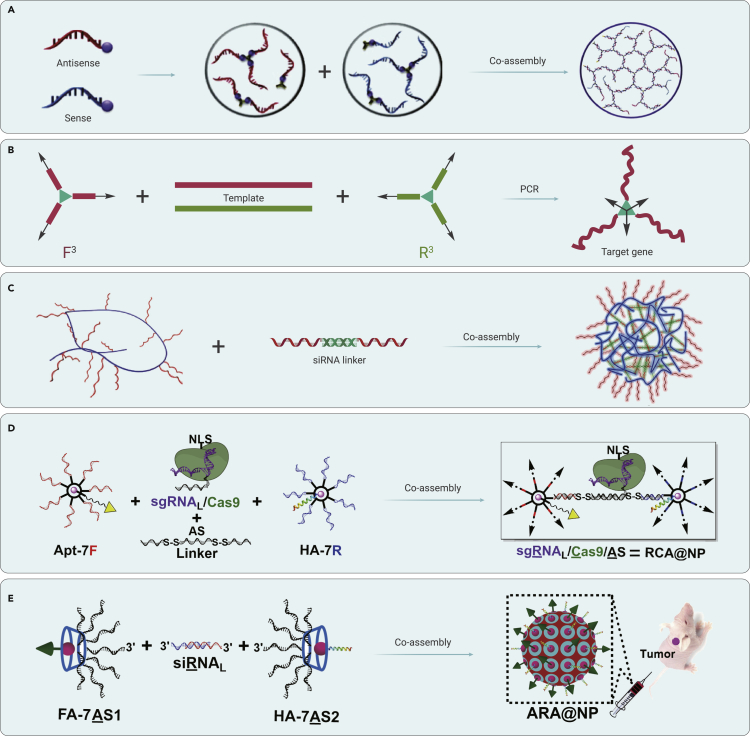
In 2011, Hong et al. employed a branched organic molecule to crosslink the 3′ terminals of sense and antisense strands of siRNA, respectively (Figure 8A).116 The covalently crosslinked RNA strands co-assembled with each other through complementary hybridization to form an siRNA-based microhydrogel. This RNA microhydrogel was efficiently condensed by a cationic polymer (LPEI) to result in a compact and stable nanoparticle. The nanoparticle demonstrated successful cellular uptake and gene-silencing effects induced by the biologically active, siRNA-based microhydrogel. This work represents another type of siRNA formulation for potential application in vivo. Inspired by this crosslinking strategy for drug delivery, in 2015, Liu et al. synthesized a branched primer pair for the efficient construction of a gene nanoparticle by branched PCR in a one-pot system (Figure 8B).117 This gene nanoparticle possessed multiple gene copies and demonstrated enhanced stability against digestion by exonuclease III. Notably, the gene nanoparticle functioned normally as a poly linear vector for protein expression in eukaryotic cells. Furthermore, in 2018, the same group constructed a gene nanoparticle encoding the p53 tumor suppressor gene, through branched PCR, for tumor therapy.118 The p53 gene nanoparticle exhibited efficient anti-tumor activity by upregulation of p53 in tumor cells. These reports describe a promising nanoplatform for the efficient construction of stable gene vectors for gene therapy.
Figure 8.
Delivery of nucleic acid drugs based on chemically modified nucleic acid nanostructures
(A) Gene silencing by siRNA microhydrogels. Reproduced with permission from Hong et al.116 Copyright 2011, American Chemical Society.
(B) Stable gene nanoparticles constructed by branched PCR for gene delivery. Reproduced with permission from Liu et al.117 Copyright 2015, Royal Society of Chemistry.
(C) Crosslinked nucleic acid nanogel for siRNA delivery. Reproduced with permission from Ding et al.119 Copyright 2018, Wiley-VCH.
(D) Branched DNA for sgRNA/Cas9/antisense delivery. Reproduced with permission from Liu et al.120 Copyright 2019, American Chemical Society.
(E) Branched antisense and siRNA co-assembled nanoparticle for combined gene silencing. Reproduced with permission from Liu et al.121 Copyright 2021, Wiley-VCH.
To precisely load nucleic acid drugs, in 2016, Bujold et al. synthesized a chemically conjugated DNA “nanosuitcase” capable of encapsulating siRNA.122 The DNA nanosuitcase protected its cargo against nuclease degradation. The loaded siRNA was stimuli-responsively released upon recognizing a nucleic acid trigger, such as endogenous mRNA or microRNA. The released siRNA and captured triggers were then responsive to exert a synergistic therapy. This smart siRNA delivery carrier was demonstrated to function in the complex cellular environment. This report provides an outstanding example of the delivery of nucleic acid drugs based on a chemically modified, wireframe DNA nanocage. Furthermore, in 2018, Ding et al. synthesized a DNA-grafted polymer to capture terminal-extended siRNA through DNA/RNA hybridization (Figure 8C).119 After co-assembly, a DNA-RNA hybrid nanogel with tunable size was constructed. This type of nanogel demonstrated enhanced resistance to enzymatic degradation. The loaded siRNA can be released from the nanogel by cleavage of endogenous RNase H to induce successful gene silencing in vitro and in vivo. Similarly, in 2019, the same group employed this DNA-grafted polymer to load sgRNA/Cas9 complex for gene editing.123 After adding DNA linkers, a nanosized hydrogel with embedded gene-editing tools was fabricated. This hydrogel also demonstrated efficient cellular uptake and subsequent gene-editing effects. Moreover, in 2021, the same group developed this DNA-grafted polymer-based strategy to deliver miRNA for glioblastoma therapy.124 The virus-mimicking membrane-coated DNA/miRNA hybrid hydrogel can reprogram microglia and macrophages from a pro-invasive M2 phenotype to an anti-tumor M1 phenotype for efficient tumor growth inhibition.
For targeting delivery and efficient endosomal escape, in 2019, Liu et al. employed an azide-modified β-cyclodextrin as a core to construct a branched DNA structure with seven arms by a click reaction for crosslinking (Figure 8D).120 The branched DNA structure co-assembled with 3′ terminal-extended sgRNA through a DNA linker (antisense with two disulfide linkages). The adamantine-conjugated aptamer (Apt-Ad for targeting) and adamantine-modified influenza hemagglutinin peptide (HA-Ad for endosomal release) were then incorporated through host-guest interaction. This multifunctional DNA nanoplatform exhibited efficient cellular uptake and endosomal escape for successful gene therapy. A remarkable inhibition of tumor growth was observed, depending on the combination of gene editing (sgRNA/Cas9, targeting DNA in the nucleus) and gene silencing (antisense, targeting mRNA in the cytoplasm). Furthermore, in 2021, the same group upgraded this nanoplatform by changing the sequences of branched DNA into antisense strands (Figure 8E).121 The branched antisense can capture siRNA with 3′ overhangs by DNA/RNA hybridization to form nucleic acid nanoparticles. After cleavage by endogenous RNase H, the branched antisense and biologically active siRNA can be efficiently released for combined gene silencing of tumor-associated genes in vitro and in vivo. These works present a general strategy for the construction of nucleic acid drug nanocarriers for efficient gene therapy in vivo.
Conclusions and outlook
Over the past decades, different chemical modifications of DNA have been developed and successfully employed for various biomedical applications. In this review, we have summarized the recent progress in the construction of chemically modified DNA nanostructures. Three representative strategies, including terminal covalent modifications, branched PCR and ligation, and direct covalent modifications onto DNA origami, were highlighted. We subsequently discussed the applications of chemically modified DNA nanostructures in the field of drug delivery. The efficient loading and delivery of therapeutic drugs, including chemotherapeutic drugs and nucleic acid drugs are discussed. Multiple functional components, such as active targeting groups and stimuli-responsive elements, can be efficiently organized in the chemically modified DNA nanostructures for targeted delivery and controlled release of loaded drugs. This kind of rationally designed drug delivery system plays a significant role in the reduction of systemic toxicity observed in traditional administration and the improvement of drug pharmacodynamics.
Although various chemically modified DNA nanostructures have been successfully employed as nanocarriers for drug delivery, some critical challenges remain for their further applications in the clinic. First, the mass-production of chemically modified DNA nanostructures should be further considered. Efficient chemical reactions with mild reaction conditions and high reaction efficiency are urgently needed to synthesize chemically modified nucleic acid on a large scale. Second, the possible immunogenicity of chemically modified DNA should be taken into consideration. Fortunately, the immune responses elicited by natural DNA are mainly triggered by the unmethylated CpG motif, which can be intentionally reduced by the design of synthetic DNA sequences. Third, efficient targeted delivery and controllable endosomal escape of the chemically modified DNA nanostructures are still needed. Endosomal escape is especially essential for nucleic acid drugs, which mainly function in the cytoplasm or nucleus after internalization into cells. Finally, the systemic pharmacokinetics of chemically modified DNA nanostructures, including the circulation, distribution, metabolism, and excretion require a more robust understanding. Chemically modified DNA nanostructures with various sizes and shapes may exhibit different systemic pharmacokinetic behaviors, which have not been evaluated in detail.
Despite the above-mentioned challenges, chemically modified DNA nanostructures have attracted much attention as smart nanocarriers for drug delivery. First, chemically modified DNA can both facilitate structural assembly and avoid the synthesis of multiple DNA strands. Second, the additional covalent bonds can be further employed for functionalization. Third, chemically modified DNA nanostructures usually demonstrate enhanced enzymatic stability, thus prolonging drug efficacy. Finally, covalent conjugation can easily incorporate various targeting and controllable release elements into chemically modified DNA nanocarriers. In particular, chemically modified DNA origami with multiple modifiable sites holds great potential to precisely co-load and efficiently co-deliver various therapeutic drugs through its protective shield to enhance nuclease resistance. We envision that these chemically modified DNA nanostructures will be widely employed in the development of drug carriers and utilized in pre-clinical and clinical studies in the near future.
Acknowledgments
This work is supported by the National Key R&D Program of China (2021YFA1200302, 2018YFA0208900), the National Natural Science Foundation of China (22025201, 22077023, and 21721002), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB36000000), the CAS Interdisciplinary Innovation Team, the Youth Innovation Promotion Association CAS, and K. C. Wong Education Foundation (GJTD-2018-03).
Author contributions
J.L. and B.D. conceived and coordinated the review. Y.W., X.L., X.W., J.L., and B.D. wrote the manuscript. Y.W., X.L., X.W., Y.L., W.T., C.Y., and J.L. drew the figures. All authors were involved in the preparation of the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Jianbing Liu, Email: liujb@nanoctr.cn.
Baoquan Ding, Email: dingbq@nanoctr.cn.
Lead contact website
References
- 1.Jiang Q., Song C., Nangreave J., et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012;134:13396–13403. doi: 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- 2.Liang H., Zhang X., Lv Y., et al. Functional DNA-containing nanomaterials: cellular applications in biosensing, imaging, and targeted therapy. Acc. Chem. Res. 2014;47:1891–1901. doi: 10.1021/ar500078f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saha S., Prakash V., Halder S., et al. A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nat. Nanotechnol. 2015;10:645–651. doi: 10.1038/nnano.2015.130. [DOI] [PubMed] [Google Scholar]
- 4.Edwardson T.G., Lau K.L., Bousmail D., et al. Transfer of molecular recognition information from DNA nanostructures to gold nanoparticles. Nat. Chem. 2016;8:162–170. doi: 10.1038/nchem.2420. [DOI] [PubMed] [Google Scholar]
- 5.Funke J.J., Dietz H. Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotechnol. 2016;11:47–52. doi: 10.1038/nnano.2015.240. [DOI] [PubMed] [Google Scholar]
- 6.Funke J.J., Ketterer P., Lieleg C., et al. Uncovering the forces between nucleosomes using DNA origami. Sci. Adv. 2016;2:e1600974. doi: 10.1126/sciadv.1600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang D., Ge Z., Im H.J., et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat. Biomed. Eng. 2018;2:865–877. doi: 10.1038/s41551-018-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S., Jiang Q., Liu S., et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018;36:258–264. doi: 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- 9.Lan X., Liu T., Wang Z., et al. DNA-guided plasmonic helix with switchable chirality. J. Am. Chem. Soc. 2018;140:11763–11770. doi: 10.1021/jacs.8b06526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Q.Q., Li H., Wang L.H., et al. DNA nanotechnology-enabled drug delivery systems. Chem. Rev. 2019;119:6459–6506. doi: 10.1021/acs.chemrev.7b00663. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Q., Liu S.L., Liu J.B., et al. Rationally designed DNA-origami nanomaterials for drug delivery in vivo. Adv. Mater. 2019;31:e1804785. doi: 10.1002/adma.201804785. [DOI] [PubMed] [Google Scholar]
- 12.Wiraja C., Zhu Y., Lio D.C.S., et al. Framework nucleic acids as programmable carrier for transdermal drug delivery. Nat. Commun. 2019;10:1147. doi: 10.1038/s41467-019-09029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu T., Liu J., Liu M., et al. A nanobody-conjugated DNA nanoplatform for targeted platinum-drug delivery. Angew. Chem. Int. Ed. 2019;58:14224–14228. doi: 10.1002/anie.201909345. [DOI] [PubMed] [Google Scholar]
- 14.Liu S., Jiang Q., Zhao X., et al. A DNA nanodevice-based vaccine for cancer immunotherapy. Nat. Mater. 2021;20:421–430. doi: 10.1038/s41563-020-0793-6. [DOI] [PubMed] [Google Scholar]
- 15.Seeman N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 16.Kallenbach N.R., Ma R.I., Seeman N.C. An immobile nucleic acid junction constructed from oligonucleotides. Nature. 1983;305:829–831. [Google Scholar]
- 17.Winfree E., Liu F., Wenzler L.A., et al. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 18.Mao C., Sun W., Shen Z., et al. A nanomechanical device based on the B-Z transition of DNA. Nature. 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 19.Yan H., Zhang X., Shen Z., et al. A robust DNA mechanical device controlled by hybridization topology. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 20.Li Y., Tseng Y.D., Kwon S.Y., et al. Controlled assembly of dendrimer-like DNA. Nat. Mater. 2004;3:38–42. doi: 10.1038/nmat1045. [DOI] [PubMed] [Google Scholar]
- 21.Zheng J., Birktoft J.J., Chen Y., et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu H., Chao J., Xiao S.J., et al. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Chandrasekaran A.R., Shen Z., et al. Paranemic crossover DNA: there and back again. Chem. Rev. 2019;119:6273–6289. doi: 10.1021/acs.chemrev.8b00207. [DOI] [PubMed] [Google Scholar]
- 24.Goodman R.P., Schaap I.A., Tardin C.F., et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 25.Chen J., Seeman N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 26.Goodman R.P., Berry R.M., Turberfield A.J. The single-step synthesis of a DNA tetrahedron. Chem. Commun. 2004:1372–1373. doi: 10.1039/b402293a. [DOI] [PubMed] [Google Scholar]
- 27.He Y., Ye T., Su M., et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 28.Rothemund P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 29.Andersen E.S., Dong M., Nielsen M.M., et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 30.Dietz H., Douglas S.M., Shih W.M. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Douglas S.M., Dietz H., Liedl T., et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han D., Pal S., Nangreave J., et al. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 33.Gerling T., Wagenbauer K.F., Neuner A.M., et al. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science. 2015;347:1446–1452. doi: 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki Y., Endo M., Sugiyama H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nat. Commun. 2015;6:8052. doi: 10.1038/ncomms9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han D., Qi X., Myhrvold C., et al. Single-stranded DNA and RNA origami. Science. 2017;358:eaao2648. doi: 10.1126/science.aao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong F., Zhang F., Liu Y., et al. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 2017;117:12584–12640. doi: 10.1021/acs.chemrev.6b00825. [DOI] [PubMed] [Google Scholar]
- 37.Yao G., Zhang F., Wang F., et al. Meta-DNA structures. Nat. Chem. 2020;12:1067–1075. doi: 10.1038/s41557-020-0539-8. [DOI] [PubMed] [Google Scholar]
- 38.Ali M.M., Li F., Zhang Z., et al. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- 39.Douglas S.M., Bachelet I., Church G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 40.Lin C., Jungmann R., Leifer A.M., et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 2012;4:832–839. doi: 10.1038/nchem.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Endo M., Sugiyama H. Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Acc. Chem. Res. 2014;47:1645–1653. doi: 10.1021/ar400299m. [DOI] [PubMed] [Google Scholar]
- 42.Jones M.R., Seeman N.C., Mirkin C.A. Programmable materials and the nature of the DNA bond. Science. 2015;347:1260901. doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- 43.Ke G., Liu M., Jiang S., et al. Directional regulation of enzyme pathways through the control of substrate channeling on a DNA origami scaffold. Angew. Chem. Int. Ed. 2016;55:7483–7486. doi: 10.1002/anie.201603183. [DOI] [PubMed] [Google Scholar]
- 44.Nickels P.C., Wünsch B., Holzmeister P., et al. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science. 2016;354:305–307. doi: 10.1126/science.aah5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seeman N.C., Sleiman H.F. DNA nanotechnology. Nat. Rev. Mater. 2017;3:17068. [Google Scholar]
- 46.Liu X., Zhang F., Jing X., et al. Complex silica composite nanomaterials templated with DNA origami. Nature. 2018;559:593–598. doi: 10.1038/s41586-018-0332-7. [DOI] [PubMed] [Google Scholar]
- 47.Chao J., Wang J., Wang F., et al. Solving mazes with single-molecule DNA navigators. Nat. Mater. 2019;18:273–279. doi: 10.1038/s41563-018-0205-3. [DOI] [PubMed] [Google Scholar]
- 48.Kwon P.S., Ren S., Kwon S.J., et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat. Chem. 2020;12:26–35. doi: 10.1038/s41557-019-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veneziano R., Moyer T.J., Stone M.B., et al. Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat. Nanotechnol. 2020;15:716–723. doi: 10.1038/s41565-020-0719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madsen M., Gothelf K.V. Chemistries for DNA nanotechnology. Chem. Rev. 2019;119:6384–6458. doi: 10.1021/acs.chemrev.8b00570. [DOI] [PubMed] [Google Scholar]
- 51.Whitfield C.J., Zhang M., Winterwerber P., et al. Functional DNA-polymer conjugates. Chem. Rev. 2021;121:11030–11084. doi: 10.1021/acs.chemrev.0c01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong Y., Yao C., Zhu Y., et al. DNA functional materials assembled from branched DNA: design, synthesis, and applications. Chem. Rev. 2020;120:9420–9481. doi: 10.1021/acs.chemrev.0c00294. [DOI] [PubMed] [Google Scholar]
- 53.Fidanza J.A., Ozaki H., McLaughlin L.W. Site-specific labeling of DNA sequences containing phosphorothioate diesters. J. Am. Chem. Soc. 1992;114:5509–5517. [Google Scholar]
- 54.Lee J.H., Wong N.Y., Tan L.H., et al. Controlled alignment of multiple proteins and nanoparticles with nanometer resolution via backbone-modified phosphorothioate DNA and bifunctional linkers. J. Am. Chem. Soc. 2010;132:8906–8908. doi: 10.1021/ja103739f. [DOI] [PubMed] [Google Scholar]
- 55.Power D.G., Kemeny N.E. The role of floxuridine in metastatic liver disease. Mol. Cancer Ther. 2009;8:1015–1025. doi: 10.1158/1535-7163.MCT-08-0709. [DOI] [PubMed] [Google Scholar]
- 56.Kim K.R., Hwang D., Kim J., et al. Streptavidin-mirror DNA tetrahedron hybrid as a platform for intracellular and tumor delivery of enzymes. J. Control Release. 2018;280:1–10. doi: 10.1016/j.jconrel.2018.04.051. [DOI] [PubMed] [Google Scholar]
- 57.Thai H.B.D., Kim K.R., Hong K.T., et al. Kidney-targeted cytosolic delivery of siRNA using a small-sized mirror DNA tetrahedron for enhanced potency. ACS Cent. Sci. 2020;6:2250–2258. doi: 10.1021/acscentsci.0c00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheffler M., Dorenbeck A., Jordan S., et al. Self-assembly of trisoligonucleotidyls: the case for nano-acetylene and nano-cyclobutadiene. Angew. Chem. Int. Ed. 1999;38:3311–3315. [PubMed] [Google Scholar]
- 59.Stepp B.R., Gibbs-Davis J.M., Koh D.L.F., et al. Cooperative melting in caged dimers of rigid small molecule-DNA hybrids. J. Am. Chem. Soc. 2008;130:9628–9629. doi: 10.1021/ja801572n. [DOI] [PubMed] [Google Scholar]
- 60.Eryazici I., Yildirim I., Schatz G.C., et al. Enhancing the melting properties of small molecule-DNA hybrids through designed hydrophobic interactions: an experimental-computational study. J. Am. Chem. Soc. 2012;134:7450–7458. doi: 10.1021/ja300322a. [DOI] [PubMed] [Google Scholar]
- 61.Hong B.J., Cho V.Y., Bleher R., et al. Enhancing DNA-mediated assemblies of supramolecular cage dimers through tuning core flexibility and DNA length-a combined experimental-modeling study. J. Am. Chem. Soc. 2015;137:13381–13388. doi: 10.1021/jacs.5b08678. [DOI] [PubMed] [Google Scholar]
- 62.Hong B.J., Eryazici I., Bleher R., et al. Directed assembly of nucleic acid-based polymeric nanoparticles from molecular tetravalent cores. J. Am. Chem. Soc. 2015;137:8184–8191. doi: 10.1021/jacs.5b03485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aldaye F.A., Sleiman H.F. Sequential self-assembly of a DNA hexagon as a template for the organization of gold nanoparticles. Angew. Chem. Int. Ed. 2006;45:2204–2209. doi: 10.1002/anie.200502481. [DOI] [PubMed] [Google Scholar]
- 64.Aldaye F.A., Sleiman H.F. Guest-mediated access to a single DNA nanostructure from a library of multiple assemblies. J. Am. Chem. Soc. 2007;129:10070–10071. doi: 10.1021/ja073305n. [DOI] [PubMed] [Google Scholar]
- 65.Aldaye F.A., Sleiman H.F. Modular access to structurally switchable 3D discrete DNA assemblies. J. Am. Chem. Soc. 2007;129:13376–13377. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- 66.Zimmermann J., Cebulla M.P.J., Mönninghoff S., et al. Self-assembly of a DNA dodecahedron from 20 trisoligonucleotides with C3h linkers. Angew. Chem. Int. Ed. 2008;47:3626–3630. doi: 10.1002/anie.200702682. [DOI] [PubMed] [Google Scholar]
- 67.Yang H., McLaughlin C.K., Aldaye F.A., et al. Metal-nucleic acid cages. Nat. Chem. 2009;1:390–396. doi: 10.1038/nchem.290. [DOI] [PubMed] [Google Scholar]
- 68.Lo P.K., Karam P., Aldaye F.A., et al. Loading and selective release of cargo in DNA nanotubes with longitudinal variation. Nat. Chem. 2010;2:319–328. doi: 10.1038/nchem.575. [DOI] [PubMed] [Google Scholar]
- 69.Trinh T., Liao C., Toader V., et al. DNA-imprinted polymer nanoparticles with monodispersity and prescribed DNA-strand patterns. Nat. Chem. 2018;10:184–192. doi: 10.1038/nchem.2893. [DOI] [PubMed] [Google Scholar]
- 70.Lee J.K., Jung Y.H., Stoltenberg R.M., et al. Synthesis of DNA-organic molecule-DNA triblock oligomers using the amide coupling reaction and their enzymatic amplification. J. Am. Chem. Soc. 2008;130:12854–12855. doi: 10.1021/ja8044458. [DOI] [PubMed] [Google Scholar]
- 71.Keller S., Wang J., Chandra M., et al. DNA polymerase-catalyzed DNA network growth. J. Am. Chem. Soc. 2008;130:13188–13189. doi: 10.1021/ja8045348. [DOI] [PubMed] [Google Scholar]
- 72.Lee J.K., Jung Y.H., Tok J.B.H., et al. Syntheses of organic molecule-DNA hybrid structures. ACS Nano. 2011;5:2067–2074. doi: 10.1021/nn1032455. [DOI] [PubMed] [Google Scholar]
- 73.Liu J., Li Y., Ma D., et al. Flexible DNA junction assisted efficient construction of stable gene nanoparticles for gene delivery. Chem. Commun. 2016;52:1953–1956. doi: 10.1039/c5cc07949g. [DOI] [PubMed] [Google Scholar]
- 74.Hartman M.R., Yang D.Y., Tran T.N.N., et al. Thermostable branched DNA nanostructures as modular primers for polymerase chain reaction. Angew. Chem. Int. Ed. 2013;52:8699–8702. doi: 10.1002/anie.201302175. [DOI] [PubMed] [Google Scholar]
- 75.Voigt N.V., Tørring T., Rotaru A., et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 2010;5:200–203. doi: 10.1038/nnano.2010.5. [DOI] [PubMed] [Google Scholar]
- 76.Tokura Y., Jiang Y., Welle A., et al. Bottom-up fabrication of nanopatterned polymers on DNA origami by in situ atom-transfer radical polymerization. Angew. Chem. Int. Ed. 2016;55:5692–5697. doi: 10.1002/anie.201511761. [DOI] [PubMed] [Google Scholar]
- 77.Gerling T., Kube M., Kick B., et al. Sequence-programmable covalent bonding of designed DNA assemblies. Sci. Adv. 2018;4:eaau1157. doi: 10.1126/sciadv.aau1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin Z., Xiong Y., Xiang S., et al. Controllable covalent-bound nanoarchitectures from DNA frames. J. Am. Chem. Soc. 2019;141:6797–6801. doi: 10.1021/jacs.9b01510. [DOI] [PubMed] [Google Scholar]
- 79.Burns J.R., Stulz E., Howorka S. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 2013;13:2351–2356. doi: 10.1021/nl304147f. [DOI] [PubMed] [Google Scholar]
- 80.Knudsen J.B., Liu L., Bank Kodal A.L., et al. Routing of individual polymers in designed patterns. Nat. Nanotechnol. 2015;10:892–898. doi: 10.1038/nnano.2015.190. [DOI] [PubMed] [Google Scholar]
- 81.Kim K.R., Kim H.Y., Lee Y.-D., et al. Self-assembled mirror DNA nanostructures for tumor-specific delivery of anticancer drugs. J. Control Release. 2016;243:121–131. doi: 10.1016/j.jconrel.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 82.Kim K.R., Kang S.J., Lee A.Y., et al. Highly tumor-specific DNA nanostructures discovered by in vivo screening of a nucleic acid cage library and their applications in tumor-targeted drug delivery. Biomaterials. 2019;195:1–12. doi: 10.1016/j.biomaterials.2018.12.026. [DOI] [PubMed] [Google Scholar]
- 83.Mou Q., Ma Y., Pan G., et al. DNA trojan horses: self-assembled floxuridine-containing DNA polyhedra for cancer therapy. Angew. Chem. Int. Ed. 2017;56:12528–12532. doi: 10.1002/anie.201706301. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J., Guo Y., Ding F., et al. A camptothecin-grafted DNA tetrahedron as a precise nanomedicine to inhibit tumor growth. Angew. Chem. Int. Ed. 2019;58:13794–13798. doi: 10.1002/anie.201907380. [DOI] [PubMed] [Google Scholar]
- 85.Märcher A., Nijenhuis M.A.D., Gothelf K.V. A wireframe DNA cube: antibody conjugate for targeted delivery of multiple copies of monomethyl auristatin. E. Angew. Chem. Int. Ed. 2021;60:21691–21696. doi: 10.1002/anie.202107221. [DOI] [PubMed] [Google Scholar]
- 86.Zhang J., Guo Y., Pan G., et al. Injectable drug-conjugated DNA hydrogel for local chemotherapy to prevent tumor recurrence. ACS Appl. Mater. Inter. 2020;12:21441–21449. doi: 10.1021/acsami.0c03360. [DOI] [PubMed] [Google Scholar]
- 87.Yang L., Sun H., Liu Y., et al. Self-assembled aptamer-grafted hyperbranched polymer nanocarrier for targeted and photoresponsive drug delivery. Angew. Chem. Int. Ed. 2018;57:17048–17052. doi: 10.1002/anie.201809753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo Y., Zhang J., Ding F., et al. Stressing the role of DNA as a drug carrier: synthesis of DNA-drug conjugates through grafting chemotherapeutics onto phosphorothioate oligonucleotides. Adv. Mater. 2019;31:1807533. doi: 10.1002/adma.201807533. [DOI] [PubMed] [Google Scholar]
- 89.Tran B.T., Kim J., Ahn D.R. Systemic delivery of aptamer-drug conjugates for cancer therapy using enzymatically generated self-assembled DNA nanoparticles. Nanoscale. 2020;12:22945–22951. doi: 10.1039/d0nr05652a. [DOI] [PubMed] [Google Scholar]
- 90.Geng Z., Wang L., Liu K., et al. Enhancing anti-PD-1 immunotherapy by nanomicelles self-assembled from multivalent aptamer drug conjugates. Angew. Chem. Int. Ed. 2021;60:15459–15465. doi: 10.1002/anie.202102631. [DOI] [PubMed] [Google Scholar]
- 91.Ren Y., Guo Y., Liu X., et al. Platinum(IV) prodrug-grafted phosphorothioate DNA and its self-assembled nanostructure for targeted drug delivery. Chem. J. Chin. Univ. 2020;41:1721–1730. [Google Scholar]
- 92.Zhu L., Guo Y., Qian Q., et al. Carrier-free delivery of precise drug-chemogene conjugates for synergistic treatment of drug-resistant cancer. Angew. Chem. Int. Ed. 2020;59:17944–17950. doi: 10.1002/anie.202006895. [DOI] [PubMed] [Google Scholar]
- 93.Fire A., Xu S., Montgomery M.K., et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 94.Elbashir S.M., Harborth J., Lendeckel W., et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 95.Jinek M., Chylinski K., Fonfara I., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cong L., Ran F.A., Cox D., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mali P., Yang L., Esvelt K.M., et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cox D.B.T., Platt R.J., Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 100.Rinaldi C., Wood M.J.A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2018;14:9–21. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 101.Cavazzana M., Bushman F.D., Miccio A., et al. Gene therapy targeting haematopoietic stem cells for inherited diseases: progress and challenges. Nat. Rev. Drug Discov. 2019;18:447–462. doi: 10.1038/s41573-019-0020-9. [DOI] [PubMed] [Google Scholar]
- 102.Slack F.J., Chinnaiyan A.M. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weng Y., Xiao H., Zhang J., et al. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019;37:801–825. doi: 10.1016/j.biotechadv.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 104.Ma C.C., Wang Z.L., Xu T., et al. The approved gene therapy drugs worldwide: from 1998 to 2019. Biotechnol. Adv. 2020;40:107502. doi: 10.1016/j.biotechadv.2019.107502. [DOI] [PubMed] [Google Scholar]
- 105.Tambuyzer E., Vandendriessche B., Austin C.P., et al. Therapies for rare diseases: therapeutic modalities, progress and challenges ahead. Nat. Rev. Drug Discov. 2020;19:93–111. doi: 10.1038/s41573-019-0049-9. [DOI] [PubMed] [Google Scholar]
- 106.Schüller V.J., Heidegger S., Sandholzer N., et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano. 2011;5:9696–9702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 107.Lee H., Lytton-Jean A.K., Chen Y., et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee J.B., Hong J., Bonner D.K., et al. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat. Mater. 2012;11:316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun W., Ji W., Hall J.M., et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed. 2015;54:12197–12201. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brunner K., Harder J., Halbach T., et al. Cell-penetrating and neurotargeting dendritic siRNA nanostructures. Angew. Chem. Int. Ed. 2015;54:1946–1949. doi: 10.1002/anie.201409803. [DOI] [PubMed] [Google Scholar]
- 111.Rahman M.A., Wang P., Zhao Z., et al. Systemic delivery of Bc12-targeting siRNA by DNA nanoparticles suppresses cancer cell growth. Angew. Chem. Int. Ed. 2017;56:16023–16027. doi: 10.1002/anie.201709485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu J., Song L., Liu S., et al. A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy. Nano Lett. 2018;18:3328–3334. doi: 10.1021/acs.nanolett.7b04812. [DOI] [PubMed] [Google Scholar]
- 113.Liu J., Song L., Liu S., et al. A tailored DNA nanoplatform for synergistic RNAi-/chemotherapy of multidrug-resistant tumors. Angew. Chem. Int. Ed. 2018;57:15486–15490. doi: 10.1002/anie.201809452. [DOI] [PubMed] [Google Scholar]
- 114.Sun W., Wang J., Hu Q., et al. CRISPR-Cas12a delivery by DNA-mediated bioresponsive editing for cholesterol regulation. Sci. Adv. 2020;6:eaba2983. doi: 10.1126/sciadv.aba2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu X., Wu T., Liu J., et al. Gene therapy based on nucleic acid nanostructure. Adv. Healthc. Mater. 2020;9:2001046. doi: 10.1002/adhm.202001046. [DOI] [PubMed] [Google Scholar]
- 116.Hong C.A., Lee S.H., Kim J.S., et al. Gene silencing by siRNA microhydrogels via polymeric nanoscale condensation. J. Am. Chem. Soc. 2011;133:13914–13917. doi: 10.1021/ja2056984. [DOI] [PubMed] [Google Scholar]
- 117.Liu J., Wang R., Ma D., et al. Efficient construction of stable gene nanoparticles through polymerase chain reaction with flexible branched primers for gene delivery. Chem. Commun. 2015;51:9208–9211. doi: 10.1039/c5cc01788b. [DOI] [PubMed] [Google Scholar]
- 118.Cheng L., Deng H., Ma D., et al. Branch-PCR constructed TP53 gene nanovector for potential cancer therapy. Chem. Commun. 2018;54:9687–9690. doi: 10.1039/c8cc05066j. [DOI] [PubMed] [Google Scholar]
- 119.Ding F., Mou Q., Ma Y., et al. A crosslinked nucleic acid nanogel for effective siRNA delivery and antitumor therapy. Angew. Chem. Int. Ed. 2018;57:3064–3068. doi: 10.1002/anie.201711242. [DOI] [PubMed] [Google Scholar]
- 120.Liu J., Wu T., Lu X., et al. A self-assembled platform based on branched DNA for sgRNA/Cas9/antisense delivery. J. Am. Chem. Soc. 2019;141:19032–19037. doi: 10.1021/jacs.9b09043. [DOI] [PubMed] [Google Scholar]
- 121.Liu J., Lu X., Wu T., et al. Branched antisense and siRNA co-assembled nanoplatform for combined gene silencing and tumor therapy. Angew. Chem. Int. Ed. 2021;60:1853–1860. doi: 10.1002/anie.202011174. [DOI] [PubMed] [Google Scholar]
- 122.Bujold K.E., Hsu J.C.C., Sleiman H.F. Optimized DNA "nanosuitcases" for encapsulation and conditional release of siRNA. J. Am. Chem. Soc. 2016;138:14030–14038. doi: 10.1021/jacs.6b08369. [DOI] [PubMed] [Google Scholar]
- 123.Ding F., Huang X., Gao X., et al. A non-cationic nucleic acid nanogel for the delivery of the CRISPR/Cas9 gene editing tool. Nanoscale. 2019;11:17211–17215. doi: 10.1039/c9nr05233j. [DOI] [PubMed] [Google Scholar]
- 124.Gao X., Li S., Ding F., et al. A virus-mimicking nucleic acid nanogel reprograms microglia and macrophages for glioblastoma therapy. Adv. Mater. 2021;33:2006116. doi: 10.1002/adma.202006116. [DOI] [PubMed] [Google Scholar]