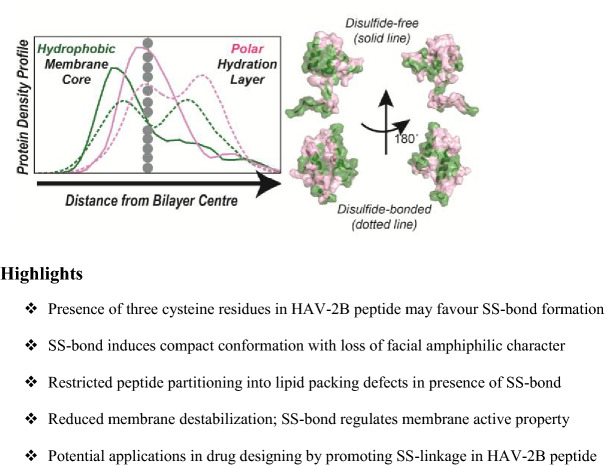
Abstract
The importance of disulphide bond in mediating viral peptide entry into host cells is well known. In the present work, we elucidate the role of disulphide (SS) bond in partitioning mechanism of membrane-active Hepatitis A Virus-2B (HAV-2B) peptide, which harbours three cysteine residues promoting formation of multiple SS-bonded states. The inclusion of SS-bond not only results in a compact conformation but also induces distorted α-helical hairpin geometry in comparison to SS-free state. Owing to these, the hydrophobic residues get buried, restricting the insertion of SS-bonded HAV-2B peptide into lipid packing defects and thus the partitioning of the peptide is completely or partly abolished. In this way, the disulphide bond can potentially regulate the partitioning of HAV-2B peptide such that the membrane remodelling effects of this viral peptide are significantly reduced. The current findings may have potential implications in drug designing, targeting the HAV-2B protein by promoting disulphide bond formation within its membrane-active region.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s00232-022-00218-0
Keywords: Disulphide bond, membrane-active viral peptide; Facial amphiphilicity; Membrane remodelling; Lipid packing defects; Molecular dynamics simulations
Highlights
Presence of three cysteine residues in HAV-2B peptide may favour SS-bond formation
SS-bond induces compact conformation with loss of facial amphiphilic character
Restricted peptide partitioning into lipid packing defects in presence of SS-bond
Reduced membrane destabilization
SS-bond regulates membrane-active property
Potential applications in drug designing by promoting SS-linkage in HAV-2B peptide
Introduction
Disulphide (SS) bond formation involves oxidation of thiol (SH) groups of two spatially proximal cysteine residues leading to a covalent linkage between their side chains. The thiol–disulphide exchange reaction, regulated by oxidoreductases, like thioredoxin and protein disulphide isomerise (PDI), (Bechtel & Weerapana 2017) predominantly occurs in endoplasmic reticulum and occasionally at other cellular sites (Wiedemann et al. 2020). The presence of disulphide bonds contributes to both protein structure and function by inducing conformational stability, facilitating protein folding and assembly and sensing changes in redox environment to modulate protein activity and localization (Bechtel & Weerapana 2017; Wiedemann et al. 2020). This plethora of functions illustrates the importance of disulphide bonds in both secretory and membrane proteins, like Prion (Jae Yoon et al. 2009; Shin et al. 2008), Src family kinases (Heppner 2021), Voltage-Dependent Anion Channel (Guardiani et al. 2016) and GPCRs (Wheatley et al. 2012).
Membrane-active peptides often harbour multiple cysteine residues, which may remain in reduced thiol state or in oxidized disulphide-linked state (Kalafatovic & Giralt 2017). Several studies have highlighted the role of redox status of cysteine residues in modulating permeability of these membrane-active peptides (Bechtel & Weerapana 2017; Fenouillet et al. 2007). For instance, antimicrobial peptides belonging to defensin family, characterized by presence of three intra-molecular SS-bridges, have been vastly studied in context of host defence mechanism against virulent microbes, by varying the number of disulphide bond and their native connectivity (Kang et al. 2019; Schroeder et al. 2011; Sharma & Nagaraj 2015; Yeasmin et al. 2021; Zhang 2020). Experiments reveal enhanced antimicrobial activity upon reduction of SS-bonds of Human defensin 1 (hBD-1) (Schroeder et al. 2011) and hBD-4 (Sharma & Nagaraj 2015). Further, molecular dynamics (MD) simulation studies of hBD-3 analogues lacking SS-linkages induce significant disruption of negatively charged lipid bilayers, compared to their native counter-part (Zhang 2020). Similarly, the antimicrobial activity of other disulphide-rich peptides has been known to be regulated by presence / absence of SS-bonds (Doherty et al. 2006; Lipkin & Lazaridis 2015; Rodnin et al. 2020; Shi et al. 2018).
Environment-dependent thiol–disulphide switching plays important role in mediating virus entry into host cells (Fenouillet et al. 2007). In this regard, it is required for Human immunodeficiency virus-1 (HIV-1) envelope protein to dissociate into two subunits, namely, gp120 and gp41, upon interaction with host cell receptors followed by reduction of redox-active SS-bonds to allosterically unmask membrane-active fusion peptide initiating membrane insertion (Ashkenazi et al. 2011; Barbouche et al. 2003; Binley James et al. 2003; Fenouillet et al. 2007; Gallina et al. 2002). Such thiol–disulphide exchange-mediated exposure and subsequent insertion of fusion peptide has been reported for other viruses as well (Abell & Brown 1993; Gallagher 1996; Jain et al. 2007; Locker & Griffiths 1999; Moyer & Nemerow 2012; Wallin et al. 2004; Zokarkar et al. 2012). On the other hand, the presence of disulphide bonds within the membrane-active region of reovirus p10 fusion-associated small transmembrane (FAST) proteins (Barry et al. 2010; Key et al. 2015) and of Ebola virus delta-peptide (He et al. 2017; Pokhrel et al. 2019) are quintessential for membrane permeation as demonstrated in recent studies.
Motivated by these studies, we investigate the role of disulphide bond in regulating membrane partitioning of Hepatitis A virus (HAV) 2B protein (Vives-Adrian et al. 2015). Compared to 2B proteins of other picornaviruses, the HAV-2B is unusually longer and shares limited (< 20%) sequence similarity with them (de Jong et al. 2008; Nieva et al. 2003). It plays a vital role in membrane remodelling (Jecht et al. 1998) and viral replication (Graff & Emerson 2003), but does not participate in calcium homeostasis or host membrane trafficking (de Jong et al. 2008). The HAV-2B protein is mainly localized in endoplasmic reticulum membrane and partly in mitochondrial, Golgi bodies and plasma membrane (Shukla et al. 2015). The membrane-active part, 60 amino acids long, located at C-terminal region of HAV-2B protein (Shukla et al. 2015) is characterized by presence of multiple cysteine residues. Experimental demonstrations, based on biophysical techniques and biochemical assays in membrane mimicking conditions, indicated an helical conformation exhibiting lipid type and composition-dependent membrane permeabilizing property (Shukla et al. 2015). In our previous study (Sikdar et al. 2021) based on extensive all-atom MD simulations, we provided insight into HAV-2B peptide-induced membrane response as a function of lipid type and composition. The simulations elucidated how the SS-free state of the peptide could sense membrane topography in the form of lipid packing defects and subsequently partitioned into model POPC bilayer, thereby inducing membrane destabilization. We also reported that presence of cholesterol significantly reduced lipid packing defects and consequently mitigated peptide recognition and partitioning into cholesterol-rich membranes. The presence of multiple cysteine residues in membrane-active region of HAV-2B and its localization preference on ER membrane, the site of thiol–disulphide exchange activity may promote formation of disulphide bonds. In the current study, we extend our earlier work by considering how chemical changes within the peptide can affect HAV-2B partitioning into model POPC membranes.
The system of our choice, HAV-2B peptide has three cysteine (C11, C47 and C52) residues. Two out of these three residues may form a SS-bond between them depending on spatial proximity, whilst the other remains free, resulting in three possible SS-linked states of HAV-2B peptide, denoted as SS11-47, SS11-52 and SS47-52. We perform 500-ns-long MD simulations in each of these SS-bonded states and compare with the SS-free HAV-2B peptide to understand how introduction of disulphide bonds may affect partitioning into POPC membrane. The SS-linkage induces shrinking of peptide conformation as well as distortion of its α-helical hairpin geometry in all three states. Further, the presence of disulphide bond sequesters hydrophobic residues, restricting insertion and partitioning of HAV-2B peptide into lipid packing defects and subsequently reducing membrane destabilization, unlike that of SS-free state. The disulphide bond thus regulates the membrane-active property of the viral peptide. The present findings may have potential implications in drug designing, targeting the HAV-2B protein by promoting disulphide bond formation within its membrane-active region. Such therapeutic applications have been implemented or under consideration for treatment of HIV and coronavirus infections (Fenouillet et al. 2007; Suhail et al. 2020).
Methods
The HAV-2B membrane-active viral peptide is 60 amino acids long and harbours three cysteine (C11, C47 and C52) residues (Fig. 1). These cysteine residues may remain in reduced thiol state or in oxidized state, where two such residues are bonded through a disulphide linkage, resulting in three possible SS-bonded states of HAV-2B peptide denoted as SS11-47, SS11-52 and SS47-52. Owing to unavailability of experimentally determined structure, the membrane-active region of HAV-2B peptide has been modelled in a recent study (Shukla et al. 2015) and further refined through extensive molecular dynamics (MD) simulations in water, details of which are provided in our earlier study (Sikdar et al. 2021). We consider representative MD snapshots of SS-free state of HAV-2B peptide in water to model the SS-bonded states. These snapshots resemble an extended state-like conformation, where a pair of cysteine residues in close proximity (~ 5 Å) can potentially form a SS-bond between them. The modelled disulphide-bonded peptides are further energy minimized over 5000 steps using the conjugate gradient algorithm in NAMD2.10 (Phillips et al. 2005). We place these energy minimized SS-bonded peptides close (~ 15 Å) to the upper leaflet of previously equilibrated 1-palmitoyl-2-oleoyl-sn-glycero-3-Phosphatidylcholine (POPC) bilayer of surface area ~ 100 × 100 Å2, comprising 147 lipid molecules per leaflet. Sufficient water molecules and counter ions are added to achieve a salt concentration of 0.15 M.
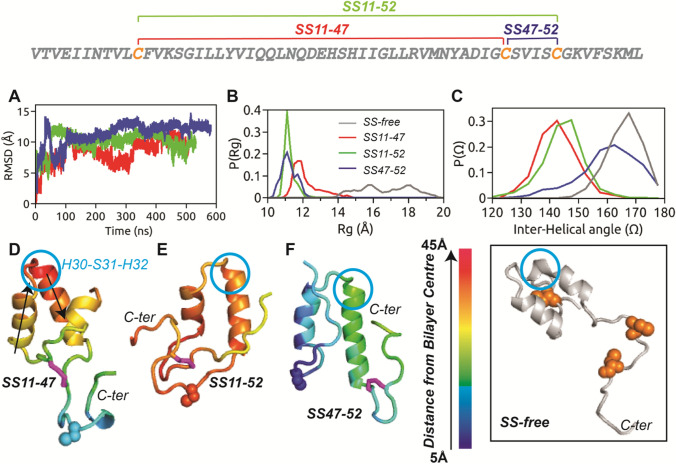
Fig. 1.
The primary sequence of HAV-2B peptide is shown with the three cysteine residues highlighted in orange, which can form the three possible SS-bonded states. a Cα-RMSD of HAV-2B peptide in different SS-bonded states with respect to corresponding initial configurations, showing flat trajectory over the last 200 ns. The equilibrium distributions of b radius of gyration, and c inter-helical angle, of disulphide-bonded states of HAV-2B peptide: SS11-47 (red), SS11-52 (green) and SS47-52 (blue). and of SS-free HAV-2B peptide (grey) are shown for comparison. The final snapshots of d SS11-47, e SS11-52 and f SS47-52 peptides are illustrated with colour codes representing insertion depth into POPC bilayer; the smaller the value, the deeper is the insertion. The SS-bond is shown in stick representation (magenta), whilst the cysteine in reduced thiol state is shown in spheres. The arrows in (d) indicate the vectors corresponding to helical axes, used for calculating the inter-helical angle, . The residue triad, H30-S31-H32 which undergoes conformational transition upon inclusion of disulphide bond is indicated by blue circle. The inset shows the largely extended conformation of SS-free HAV-2B peptide and the position of cysteine residues (spheres in orange)
MD simulations for each of these systems (see Supplementary Information, SI Table S1, Fig. S1a-c) are performed using NAMD2.10 (Phillips et al. 2005) and modified TIP3P (Jorgensen et al. 1983) water model, CHARMM36m (Huang et al. 2017) and CHARMM36 (Klauda et al. 2010) force field parameters for the peptide and the lipid molecules, respectively. All systems are energy minimized for 10,000 steps. The simulations are carried out using periodic boundary conditions in NPT ensemble at 1 atm pressure and 310 K with a time step of 2 femtoseconds. The van der Waals interactions are smoothly truncated beyond 12 Å, by a force-based switching function between 10 Å and 12 Å, whilst Particle mesh Ewald fast Fourier transform is used for electrostatic interactions. The peptide heavy atoms are additionally subjected to positional restraints, which are gradually decreased over six cycles of equilibration of 500 ps each to ensure relaxed starting configurations of SS-bonded states. The production runs are performed for over 500 ns and further analysis on equilibrated trajectories is executed using Visual Molecular Dynamics (VMD) (Humphrey et al. 1996), MEMBPLUGIN (Guixà-González et al. 2014), Packmem (Gautier et al. 2018) and in-house Fortran codes.
Results
The MD simulation of disulphide-free HAV-2B peptide in water from our previous study (Sikdar et al. 2021) indicates close spatial proximity of cysteine residues, namely C11, C47 and C52, which may favour formation of possible SS-bonded states of the peptide denoted as SS11-47, SS11-52 and SS47-52. The distributions of pairwise SS-distance of disulphide-free HAV-2B peptide in water (see SI, Fig. S2) are multi-modal with several peaks spread between 3 Å and 20 Å. Relevant MD snapshots with SS-distance < 5 Å are chosen to build respective SS-bonded states, energy minimized and further subjected to restrained MD equilibration protocol as described in the Methods section. We perform 500-ns-long all-atom MD simulations for each of these states in model POPC membrane. The root mean square deviation (RMSD) plots in Fig. 1a based on C-α atoms with respect to corresponding initial configurations show structural re-arrangements within the first 300 ns, which we consider as the equilibration period of the SS-bonded peptides in POPC bilayer. The RMSD being flat beyond 300 ns, further analysis is performed on the equilibrated trajectories of last 200 ns. We first present the conformational preferences and the membrane binding mode of different disulphide-bonded states. We follow this up with SS-bonded peptide-induced membrane response and the role of interfacial packing defects in peptide partitioning. Finally, we elucidate how introduction of the disulphide bond may regulate membrane-active property of HAV-2B peptide.
Conformational Preferences
The molecular dimension of HAV-2B peptide is characterized by probability distribution of radius of gyration, in Fig. 1b, computed as the average distance of C-α atoms from their centre of mass over the equilibrated trajectories. The SS11-47 and SS47-52 peptides explore conformational states with varying between 11 Å and 14 Å, resulting in broad , in contrast to the sharp distribution of SS11-52 peptide, with ~ 11 Å. The presence of SS-bond results in compact conformations compared to the largely extended SS-free state of HAV-2B peptide (see inset Fig. 1) in POPC membrane as indicated by in Fig. 1b. The final snapshots illustrating the conformational preferences of the three SS-bonded states of HAV-2B peptide shown in Fig. 1d-f also indicate the location of disulphide bonds. For instance, the SS-linkage connects the N- and the C-terminal tails in SS11-47 (Fig. 1d) and SS11-52 (Fig. 1e) peptides, whilst, the linkage is confined within the C-terminal tail of SS47-52 peptide. Despite the differences in location and connectivity, these disulphide bonds act as a constraint reducing the overall conformational fluctuations of the peptide.
We also characterize the conformational preference in terms of the helical arrangement of the hairpin structure through inter-helical angle, . The vector between C-α atoms of L18 and I22 represents the first helical axis, whilst that of L36 and M40 represents the second, being the angle between them (see Fig. 1d). The equilibrium distributions, of the SS-bonded states of HAV-2B peptide are shown in Fig. 1c. of SS11-47 and SS11-52 peptides are overlapping with lying between 140° and 150°, resembling a “boomerang” conformation. In contrast, the distribution of inter-helical angle of SS47-52 peptide about ~ 160° is quite similar to that of SS-free state ( ~ 170°) resembling nearly an anti-parallel helical conformation characteristic of a hairpin structure. The SS-linkage between the two terminals thus controls the inter-helical angle in a way so as to distort the hairpin geometry of both SS11-47 and SS11-52 states; whereas, the geometry is almost preserved when the linkage is confined within the C-terminal tail of SS47-52 peptide.
The secondary structure of disulphide-bonded HAV-2B peptides are calculated using the STRIDE (Frishman & Argos 1995) algorithm implemented in VMD (Humphrey et al. 1996). The residue-wise secondary structure percentage (see SI, Fig. S3) represents the population of different structural elements explored by each residue during the course of simulation. The residues I17 to L25 and H30 to Y42 form the two -helices of the hairpin motif of the SS-bonded states. The overall secondary structure percentage remains qualitatively similar across different SS-bonded states. The N- and C-terminal tail residues are predominantly characterized by “turn” or random “coil” conformations, with few residues showing minor populations of -helix, -sheets and -strands.
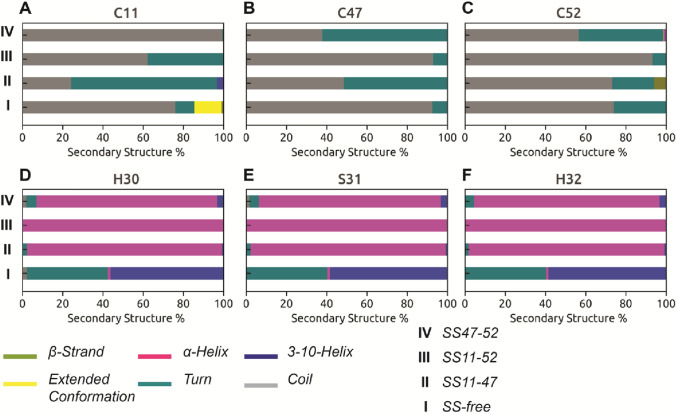
We also compare the changes in secondary structure population of the cysteine residues in different SS-bonded states (Fig. 2a-c) with that of SS-free state. C11 (Fig. 2a) predominantly exhibits random “coil” conformation in SS-free as well as in SS47-52 states. But whenever, C11 is involved in SS-bond formation as in SS11-47 and SS11-52 states, the propensity to adopt random “coil” conformation decreases with an increase in “turn”-like secondary structural element. Similarly, random “coil” conformation of C47 (Fig. 2b) is prevalent in SS-free and SS11-52 states, but enhanced population of “turn” is observed in SS11-47 and SS47-52 states. Our observations indicate that the cysteine residues upon being involved in SS-bonding generally show a shift in conformational equilibrium towards “turn”-like secondary structure component from random “coil” conformation, the only exception being increased “coil” propensity for C52 (Fig. 2c) in SS11-52 state. The presence of disulphide bond significantly alters the secondary structure propensity of a triad of residues, namely, H30-S31-H32 (see Fig. 2d-f), and the location of this triad is indicated in Fig. 1d-f. The residue triad forms part of the inter-helical flexible linker in SS-free state and shows transition between “turn” and -helices. In contrast, these residues adopt an -helical conformation all throughout the SS-bonded states (Fig. 2d-f). The change in conformational equilibrium of H30-S31-H32 from flexible “turn” / -helical to more rigid -helical conformation possibly accounts for the observed deviation in inter-helical angles (Fig. 1c) of SS-bonded states from that of SS-free state.
Fig. 2.
Percentage population of different secondary structural elements accessible to a C11, b C47, c C52, d H30, e S31 and f H32 in SS-bonded and -free states of HAV-2B peptide
Peptide–Bilayer Mode of Binding
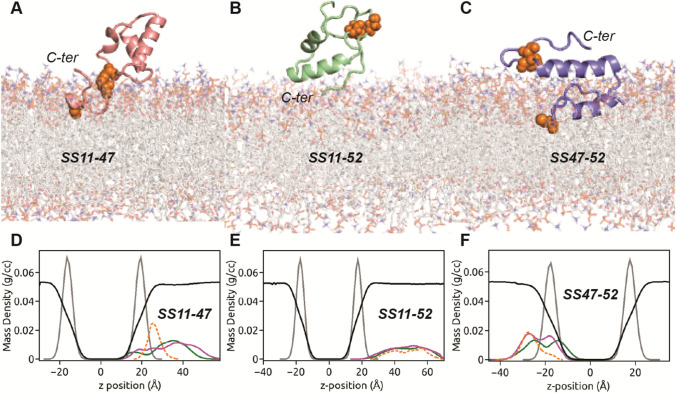
We observe significant differences in mode of peptide binding to POPC bilayer depending on disulphide connectivity (Fig. 3). The SS11-47 peptide interacts with POPC membrane (Fig. 3a) through its C-terminal region. The α-helical hairpin motif orients itself almost parallel to membrane normal (z-direction) and remains solvent exposed, including the N-terminal tail. The localization of peptide residues on bilayer is quantified through its insertion depth, calculated as the distance of centre of mass of each peptide residue from bilayer centre along the z-direction. The final snapshots of the SS-bonded peptides in Fig. 1d-f are colour coded according to residue insertion depths: The smaller the value, the deeper is the insertion. Figure 1d indicates that the C-terminal residue stretch V49 – F56 of SS11-47 peptide is located in close proximity (~ 15 Å) to bilayer centre indicating insertion, in contrast to the α-helical hairpin motif and the N-terminal tail, which remain far (~ 40 Å) from membrane interior. The SS11-52 peptide hovers close to membrane surface but fails to form any stable contacts with POPC membrane (Fig. 3b) within our simulation time scale of 500 ns. As a consequence, the peptide residues remain far (~ 40 Å) from bilayer centre (Fig. 1e). The binding mode of SS47-52 peptide with POPC membrane is shown in Fig. 3c. Unlike SS11-47 peptide, the α-helical hairpin motif orients itself parallel to bilayer resulting in enhanced contact surface area. The SS47-52 peptide interacts with the membrane by complete insertion of N-terminal helix (I17 – L25) and the preceding tail, the average insertion depth being ~ 10 – 15 Å (Fig. 1f). The C-terminal helix (H30 – Y42) of the hairpin motif and the succeeding tail region, although solvent exposed, is located close to the POPC headgroups.
Fig. 3.
The binding modes of a SS11-47, b SS11-52 and c SS47-52 peptides with POPC bilayer are shown. The cysteine (both SS-linked and free) residues are represented as orange spheres, whilst hydrating water is not shown for clarity. The mass density profiles along the bilayer normal (z-direction) of d SS11-47, e SS11-52 and f SS47-52 peptides in POPC bilayer. The density profiles comprising hydrophobic (green) and hydrophilic (magenta), SS-bonded cysteine (orange) residues of peptide, phosphate headgroups (grey) and water (reduced by factor of 10, black) are indicated
The mode of binding influences the degree of membrane-induced partitioning of peptide. The partitioning of SS11-47 peptide in POPC bilayer is illustrated through density profile of peptide atoms along the membrane normal (z-direction) as a function of distance from membrane centre (Fig. 3d), calculated over the equilibrated trajectory. As bulk of the SS11-47 peptide remains in water, the density profiles of constituent hydrophobic and hydrophilic residues are overlapping with peak positions in solvent. The peptide density profiles also show little overlap with lipid headgroups owing to insertion of few C-terminal residues. Since the SS11-52 peptide fails to insert into membrane, the corresponding density profile in Fig. 3e is entirely positioned in solvent, rarely overlapping with lipid headgroups. Although, we initially place the SS47-52 peptide close to the top leaflet, during the course of simulation, the peptide leaves the central simulation box from the top to re-enter from the bottom and localizes on the lower leaflet. This results in the density profile of SS47-52 peptide to overlap with that of lipid headgroups from the lower leaflet, as indicated in Fig. 3f. The SS47-52 peptide partitions into the membrane milieu with segregation of hydrophobic and hydrophilic density peaks toward bilayer centre and headgroups, respectively, acquiring partly facially amphiphilic conformation in POPC bilayer. Both hydrophobic and hydrophilic density profiles being bimodal, a second peak is observed in solvent close to the headgroup–water interface. This bimodal nature of density profiles is attributed to the observed horizontal binding mode of SS47-52 (Fig. 3c), characterized by membrane-embedded N-terminal helix and surface-adsorbed C-terminal helix near the POPC headgroups.
The horizontal orientation of SS47-52 peptide, parallel to membrane surface, is quite similar to the observed binding mode of SS-free HAV-2B peptide, except that in the latter both helices of the hairpin motif and the long-stretched C-terminal tail remain more deeply (~ 5–10 Å from bilayer centre) embedded within the POPC membrane (Sikdar et al. 2021). Thus more number of hydrophobic and hydrophilic residues form extensive contacts with lipid molecules resulting in enhanced density profile peak intensities of SS-free state (see SI, Fig. S4) compared to SS47-52 peptide. Further, the segregation of hydrophobic and hydrophilic peak intensities facilitates the SS-free HAV-2B peptide to acquire a strong facially amphiphilic conformation in POPC membrane. Our results strongly demonstrate that disulphide connectivity can regulate the extent of HAV-2B peptide partitioning. The disulphide linkage connecting the N- and C-terminal tails of HAV-2B peptide, as in SS11-47 and SS11-52 states, mitigates peptide partitioning. Whereas, the SS-linkage confined within the C-terminal tail involving the C47-C52 pair results in peptide partitioning, albeit weaker than the SS-free state.
In this regard, it is also interesting to study the partitioning of cysteine residues. The density peaks of disulphide-bonded cysteine pairs in SS11-47 (Fig. 3d) and SS47-52 (Fig. 3f) states are located at the solvent proximal interface very close to the lipid headgroups. Whilst the other cysteine in reduced thiol state: C52 of SS11-47 (Fig. 3a) and C11 of SS47-52 (Fig. 3c) peptides remain embedded in the hydrophobic membrane core. This is in agreement to a recent study, which concluded that cysteine residues in reduced thiol state favourably partitions into the hydrophobic membrane milieu rather than at the polar lipid–water interface (Iyer & Mahalakshmi 2019).
Influences on Membrane Properties
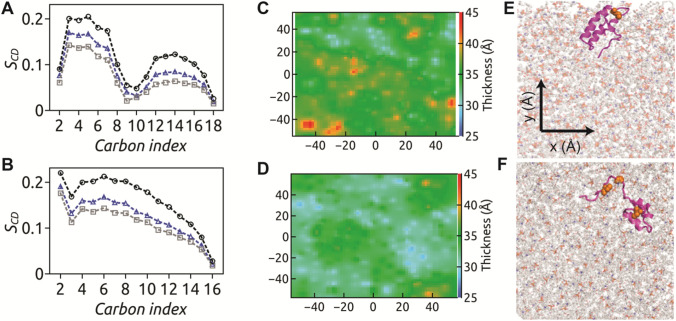
We investigate how membrane properties are affected upon partitioning of SS47-52 peptide in model POPC membrane. To this end, we quantify the flexibility of lipid acyl chains through lipid tail order parameter, , being the angle between C–H bond vector and bilayer normal, computed over the equilibrated trajectories for both saturated () and unsaturated () acyl chain carbon atoms and shown in Fig. 4. Higher the order parameter, the lower is the flexibility of lipid acyl chains and vice versa. The order parameter values, of (Fig. 4a) and (Fig. 4b) chains of POPC under the influence of SS47-52 peptide partitioning are intermediate between that of control POPC bilayer (without any peptide) and in presence of SS-free HAV-2B peptide. The presence of C47-C52 disulphide linkage enhances the acyl chain flexibility compared to control POPC bilayer; however, the disordering effects are not as strong as in the presence of the SS-free peptide. The enhanced flexibility of lipid tails causes lateral expansion of bilayer, leading to increased surface area-per-lipid (SA/lipid) and reduced bilayer thickness. The effect of SS47-52 peptide on SA/lipid (~ 74 Å2) and bilayer thickness (~ 35.6 Å) is also observed to be intermediate between that of SS-free peptide and control POPC bilayers (Sikdar et al. 2021). We illustrate the 2-d thickness profiles (Fig. 4c, d) along the membrane xy-plane corresponding to final snapshots (Fig. 4e, f) of SS47-52 and SS-free peptide–membrane systems. The lateral thickness profile reflects membrane thinning localized around the SS47-52 peptide, with thickness varying between ~ 30 Å at insertion site and ~ 40 Å elsewhere. In contrast, the SS-free state of HAV-2B peptide induces uniform global thinning of POPC bilayer, the effect being more pronounced (~ 30–35 Å) compared to the SS47-52 peptide. Owing to the reduced partitioning of HAV-2B peptide in presence of SS-bond, the membrane perturbation effects are also significantly reduced.
Fig. 4.
The order parameter, of a and b acyl chains of POPC corresponding to SS47-52 (blue triangle), SS-free peptide (grey square) and control POPC (black circle) systems are shown. The membrane thickness maps upon interaction of c SS47-52 and d SS-free states of HAV-2B peptide with POPC bilayer are generated considering inter-leaflet P–P distance with 2 Å resolution along the xy-plane. The localization of e SS47-52 and f SS-free peptide on POPC bilayer are shown. The membrane thinning effect is localized around the insertion site of SS47-52 peptide, whilst more pronounced uniform global thinning effect is observed upon SS-free HAV-2B peptide partitioning
Distribution of Lipid Packing Defects
The membrane response is not only restricted to lipid acyl tails but also extended to the bilayer–water interface in the form of lipid headgroups packing. This interfacial region is characterized by transient exposure of membrane hydrophobic core to hydration layer, leading to lipid packing defects which act as binding hotspots for peptides (Baul & Vemparala 2017; Cui et al. 2011; Garten et al. 2015; Ouberai et al. 2013; Pinot et al. 2014; Read et al. 2015; Vanni et al. 2014, 2019, 2013; Wildermuth et al. 2019). These lipid packing defects are qualitatively characterized into “Deep” or “Shallow” depending on the relative depth of the defect site with respect to the nearest glycerol backbone and further quantified by area () of the defect site, following the standard protocol using Packmem (Gautier et al. 2018).
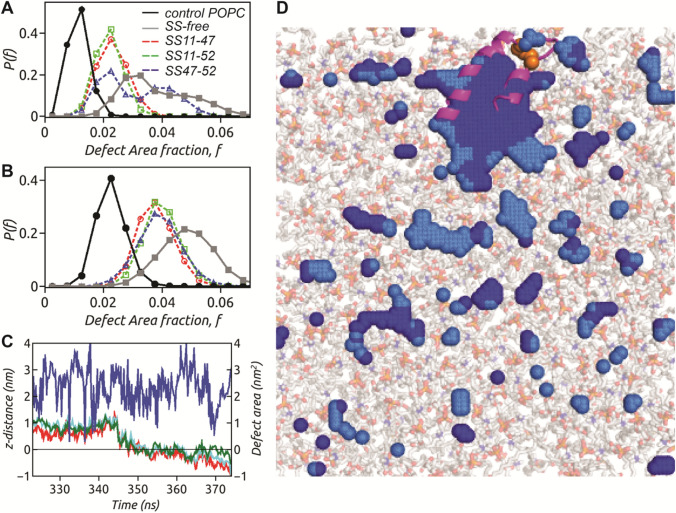
In order to quantify the extent of defects in a given frame, we calculate the total defect area in a given frame by adding the individual areas of all “Deep” (or “Shallow”) defect sites in the frame and normalize by the area of leaflet, to define the “Deep” (or “Shallow”) defect area fraction (or ) in the given frame. It provides a measure of how much leaflet area is covered by “Deep” (or “Shallow”) lipid packing defects in a given frame. The distribution of “Deep” defect area fraction, due to presence of SS-bonded peptides in POPC bilayer is shown in Fig. 5a. of SS11-47 and SS11-52 are single peaked around defect area fractional values ~ 0.02. The overlapping distributions indicate that the extent of “Deep” defects is similar for both these “non-partitioning” SS-bonded peptides but more compared to the control system. On contrary, of SS47-52 and SS-free state are broad and have significant overlap at higher defect area fraction values, ~ 0.04. This implies that large amount of leaflet area is covered by “Deep” defects in presence of these peptides. Since both the helices of SS-free peptide partition into POPC bilayer, the defect area fraction is slightly higher compared to SS47-52, where only the N-terminal helix partitions. The extent of “Shallow” defects, similar in all SS-bonded systems due to overlapping (Fig. 5b) with peak around 0.04, is intermediate between that of control POPC and SS-free system. Unlike the “Deep” defects, the “Shallow” defects remain unaffected by partitioning of SS47-52 peptide. Whilst “Deep” defects are affected by partitioning of bulky hydrophobic residues, “Shallow” defects are known to be influenced by presence of short-chain hydrophobic amino acids, as reported for Synuclein (Garten et al. 2015; Pinot et al. 2014). The N-terminal helix of hairpin motif and the preceding tail harbours few such small hydrophobic residues, whilst the majority of them reside at the C-terminal helix and the succeeding tail. Owing to this, the partitioning of N-terminal helix of SS47-52 does not affect the “Shallow” defects. On contrary, both the helices and the C-terminal tail of SS-free peptide being involved in membrane partitioning enhance the “Shallow” defects.
Fig. 5.
The distribution of defect area fraction corresponding to a “Deep” and b “Shallow” provide insight into the extent of defects in a given frame relative to leaflet area. The open symbols with dotted line represent SS11-47 (red circle), SS11-52 (green square) and SS47-52 (blue triangle) systems, whilst solid symbols with solid lines indicate SS-free peptide (grey square) and control (black circle) POPC system. c The insertion dynamics of C11 (red), L18 (cyan) and L25 (green) of SS47-52 peptide into a co-localized “Deep” defect area (blue). d The final snapshot of SS47-52 peptide with its N-terminal helix completely embedded into the large co-localized “Deep” defect (dark blue) surrounded by “Shallow” defects (light blue). The SS-linked cysteine residues (orange spheres) fail to insert into the co-localized defect
Sensing of Lipid Packing Defects
Our previous study indicated that the SS-free HAV-2B peptide senses membrane topography in the form of lipid packing defects, inserts into such defects and subsequently partitions into POPC bilayer (Sikdar et al. 2021). In this section, we elucidate the mechanism of partitioning of SS47-52 state of the viral peptide. We consider a representative set of residues, C11 (red), L18 (cyan) and L25 (green) of SS47-52 peptide (Fig. 5c), which undergo deep insertion into POPC membrane. The insertion dynamics of these residues are monitored from the individual distance (z-distance) of residue centre of mass from the average level of C2 atoms of glycerol moieties in POPC molecule, along the z-direction. A negative value of z-distance implies insertion below the average C2 level. Simultaneously, we track the appearance of any underlying “Deep” lipid packing defect that is co-localized with these residues. In the process, we identify a single large co-localized “Deep” defect in vicinity of these residues. The defect area fluctuating around 250 Å2 drives the residue insertions at around 350 ns, following which the co-localized “Deep” defect area momentarily increases to 400 Å2 to accommodate the bulky hydrophobic side chains. The insertion of SS47-52 peptide into the co-localized “Deep” defect is illustrated through a representative snapshot in Fig. 5d. The horizontal orientation of the inserted N-terminal helix of the hairpin motif stabilizes the large “Deep” defect ~ 270 Å2. This single large defect contributes to the SS47-52 peptide-induced enhanced “Deep” defect area fraction in Fig. 5a. These discrete residue insertion events following the appearance of co-localized defects suggest sensing of lipid packing defects similar to the SS-free peptide.
Discussion
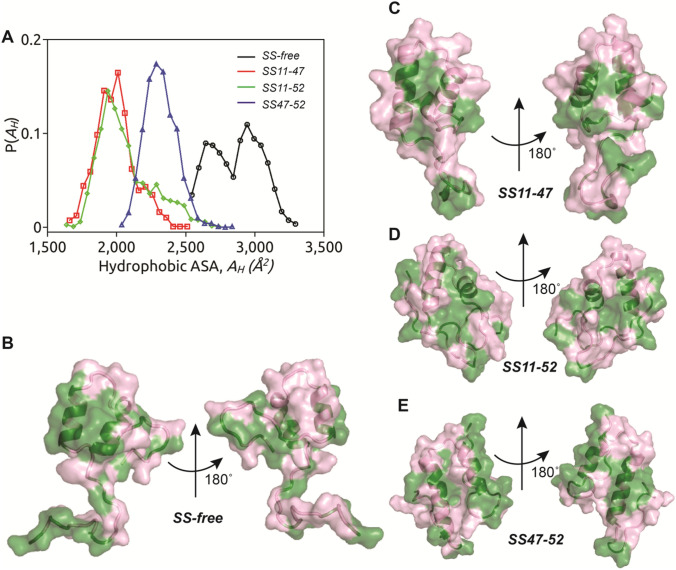
The present study provides insight into the effect of disulphide bond on HAV-2B peptide structure and partitioning into membrane. All the SS-bonded states, SS11-47, SS11-52 and SS47-52 undergo conformational shrinking, leading to compact conformations. The inclusion of SS-bond not only results in a compact conformation but also changes the inter-helical angle, , resulting in deviation from hairpin conformation in comparison to SS-free state. The anti-parallel hairpin arrangement of α-helices is known to be essential for membrane partitioning of viral peptide as reported for Influenza virus hemagglutinin (Worch et al. 2018) and Ebola virus delta-peptide (Pokhrel et al. 2019). The change in inter-helical angle, which in turn modulates helix packing interactions along with the compact conformation of SS-bonded states controls the accessible surface area (ASA) of the peptide (Fig. 6a). The hydrophobic residues being crucial in regulating HAV-2B peptide partitioning, we calculate the contribution of the same to ASA in presence and absence of the SS-bond. The hydrophobic accessible surface area () is significantly high (~ 3000 Å2) in SS-free state, followed by SS47-52 state (2300 Å2). The disulphide bond interconnecting the N- and C-terminal tails further reduces the hydrophobic exposure with < 2000 Å2 observed in SS11-47 and SS11-52 peptides, which thus fail to partition into POPC bilayer. The SS-free state of HAV-2B peptide with maximum hydrophobic exposure partitions deep into membrane milieu, whilst the partitioning ability is partly compromised with reduced hydrophobic exposure upon inclusion of SS-bond between C-terminal cysteines, C47-C52.
Fig. 6.
a The distribution of hydrophobic accessible surface area (ASA), is shown for SS-free (black), SS11-47 (red), SS11-52 (green) and SS47-52 (blue) states of HAV-2B peptide. The solvent-exposed hydrophobic (green) and hydrophilic (magenta) surface area are illustrated on the 3-D structure of HAV-2B peptide in b SS-free, c SS11-47, d SS11-52 and e SS47-52 states
We illustrate the hydrophobic (green) and hydrophilic (magenta) ASA of HAV-2B peptide in SS-free (Fig. 6b) and bound states (Fig. 6c-e). In SS-free state, the peptide presents an exposed hydrophobic-dominated face and a hydrophilic-dominated opposite surface (Fig. 6b). The conformation of SS-free HAV-2B peptide is such that it acquires a strong facially amphipihilic character upon segregation of hydrophobic and hydrophilic residues. Different membrane-active agents including antimicrobial peptides (Leontiadou et al. 2006; Mondal et al. 2010; Vanni et al. 2014), polymers (Baul et al. 2014; Baul & Vemparala, 2015, 2017; Palermo et al. 2012, 2013; Rahman et al. 2018; Rani et al. 2021) and other membrane-active molecules (Devanand et al. 2019; Polley & Vemparala, 2013; Vemparala et al. 2006) are known to acquire such amphipihilic conformations upon partitioning into cellular membranes. However, in SS11-47 (Fig. 6c) and SS11-52 (Fig. 6d) states, the hydrophobic accessible surface area is limited resulting in mitigation of peptide partitioning. On other hand, the segregation of hydrophobic and hydrophilic surfaces is not as discrete as that in SS-free state, leading to partial partitioning of SS47-52 peptide (Fig. 6e). The disulphide bond induced conformational changes thus controlling the exposure of hydrophobic residues, which in turn regulates HAV-2B peptide partitioning. As a consequence the SS-bonded peptide-induced membrane responses, in terms of lipid tail disordering, membrane thinning and abundance of interfacial packing defects, are mild compared to the SS-free state. In this way, the disulphide bond regulates the membrane-active property of HAV-2B peptide such that the membrane destabilizing effects of this viral peptide are significantly reduced.
In order to ascertain sufficient conformational sampling of the three SS-linked states, a second set of simulations is performed with different initial configurations and orientation on POPC bilayer surface (see SI, Fig. S1d-f). This second set of simulations is initiated considering compact SS-free HAV-2B peptide conformations in water. The SS-linked states remain in compact conformation all throughout the simulation time. values (see SI, Fig. S1g-i) from the two sets of simulations are similar, indicating adequate conformational sampling, irrespective of differences in starting configurations. Moreover, both sets of simulations confirm that the SS11-47 and the SS11-52 states fail to partition into the POPC bilayer. The second set of simulations of the other disulphide-bonded state, SS47-52, is run for about 200 ns to ensure that the peptide indeed represents a compact conformation irrespective of the initial setup. The results are thus consistent across both sets of simulations.
Evidence of thiol–disulphide redox status-dependent exposure of hydrophobic patches facilitating peptide partitioning is well documented through experimental investigations on different membrane-active agents, like antimicrobial peptides (Schroeder et al. 2011; Sharma & Nagaraj 2015; Shi et al. 2018; Zhang 2020) and viral peptides (Abell & Brown, 1993; Binley James et al. 2003; Gallagher 1996; Jain et al. 2007; Key et al. 2015; Locker & Griffiths 1999; Moyer & Nemerow 2012; Wallin et al. 2004; Zokarkar et al. 2012). This importance of disulphide bond in mediating viral peptide partitioning and subsequent entry into host cells is currently being explored to design antiviral agents. For instance, reduction of disulphide bond by PDI being pre-requisite for HIV entry, designing inhibitors targeting this process interferes with the virus / cell fusion mechanism (Barbouche et al. 2003; Binley James et al. 2003; Fenouillet et al. 2007; Gallina et al. 2002). Similar efforts are in progress to develop therapeutics against coronavirus infection (Suhail et al. 2020). The present findings indicate that promoting disulphide bond formation within the membrane-active HAV-2B peptide may have potential implications in designing antiviral agents to combat HAV infection.
Conclusion
The presence of multiple cysteine residues in the membrane-active region of HAV-2B peptide indicates the possibility of three SS-bonded states of the peptide. In the present work, we elucidate the role of disulphide bond in HAV-2B peptide partitioning. The SS-linkage induces shrinking of peptide conformation as well as distortion of its α-helical hairpin geometry, resulting in reduced hydrophobic exposure. Depending on disulphide connectivity, the partitioning of HAV-2B peptide is completely or partly abolished and subsequently reduced membrane remodelling effects are observed in comparison to SS-free state. The disulphide bond thus regulates the membrane-active property of the viral peptide. These results may find potential applications in drug designing approaches against HAV infection.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
All simulations in this work have been carried out on supercomputing facility Nandadevi cluster at The Institute of Mathematical Sciences, Chennai, India.
Author Contributions
SS, MB and SV designed the project. SS performed the simulations and carried out the analysis. All authors contributed to writing and reviewing of the manuscript.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/10/2022
A Correction to this paper has been published: 10.1007/s00232-022-00246-w
Contributor Information
Samapan Sikdar, Email: samapans@imsc.res.in.
Manidipa Banerjee, Email: mbanerjee@bioschool.iitd.ac.in.
Satyavani Vemparala, Email: vani@imsc.res.in.
References
- Abell BA, Brown DT. Sindbis virus membrane fusion is mediated by reduction of glycoprotein disulfide bridges at the cell surface. J Virol. 1993;67:5496–5501. doi: 10.1128/jvi.67.9.5496-5501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A, Viard M, Wexler-Cohen Y, Blumenthal R, Shai Y. Viral envelope protein folding and membrane hemifusion are enhanced by the conserved loop region of HIV-1 gp41. FASEB J. 2011;25:2156–2166. doi: 10.1096/fj.10-175752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbouche R, Miquelis R, Jones IM, Fenouillet E. Protein-disulfide Isomerase-mediated Reduction of Two Disulfide Bonds of HIV Envelope Glycoprotein 120 Occurs Post-CXCR4 Binding and Is Required for Fusion *. J Biol Chem. 2003;278:3131–3136. doi: 10.1074/jbc.M205467200. [DOI] [PubMed] [Google Scholar]
- Barry C, Key T, Haddad R, Duncan R. Features of a spatially constrained cystine loop in the p10 FAST protein ectodomain define a new class of viral fusion peptides*. J Biol Chem. 2010;285:16424–16433. doi: 10.1074/jbc.M110.118232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baul U, Vemparala S. Chapter four - membrane-bound conformations of antimicrobial agents and their modes of action. In: Iglič A, Kulkarni CV, Rappolt M, editors. Advances in planar lipid bilayers and liposomes. NY: Academic Press; 2015. pp. 97–128. [Google Scholar]
- Baul U, Vemparala S. Influence of lipid composition of model membranes on methacrylate antimicrobial polymer-membrane interactions. Soft Matter. 2017;13:7665–7676. doi: 10.1039/C7SM01211J. [DOI] [PubMed] [Google Scholar]
- Baul U, Kuroda K, Vemparala S. Interaction of multiple biomimetic antimicrobial polymers with model bacterial membranes. J Chem Phys. 2014;141:084902. doi: 10.1063/1.4893440. [DOI] [PubMed] [Google Scholar]
- Bechtel TJ, Weerapana E. From structure to redox: The diverse functional roles of disulfides and implications in disease. Proteomics. 2017 doi: 10.1002/pmic.201600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley James M, Cayanan Charmagne S, Wiley C, Schülke N, Olson William C, Burton Dennis R. Redox-triggered infection by Disulfide-Shackled Human Immunodeficiency Virus type 1 Pseudovirions. J Virol. 2003;77:5678–5684. doi: 10.1128/JVI.77.10.5678-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Lyman E, Voth GA. Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophys J. 2011;100:1271–1279. doi: 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong AS, de Mattia F, Van Dommelen MM, Lanke K, Melchers WJ, Willems PH, van Kuppeveld FJ. Functional analysis of picornavirus 2B proteins: effects on calcium homeostasis and intracellular protein trafficking. J Virol. 2008;82:3782–3790. doi: 10.1128/JVI.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanand T, Krishnaswamy S, Vemparala S. Interdigitation of lipids induced by membrane-active proteins. J Membr Biol. 2019;252:331–342. doi: 10.1007/s00232-019-00072-7. [DOI] [PubMed] [Google Scholar]
- Doherty T, Waring AJ, Hong M. Peptide–lipid interactions of the β-hairpin antimicrobial peptide tachyplesin and its linear derivatives from solid-state NMR. Biochimica Et Biophysica Acta BBA- Biomembranes. 2006;1758:1285–1291. doi: 10.1016/j.bbamem.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Fenouillet E, Barbouche R, Jones IM. Cell entry by enveloped viruses: redox considerations for HIV and SARS-coronavirus. Antioxid Redox Signal. 2007;9:1009–1034. doi: 10.1089/ars.2007.1639. [DOI] [PubMed] [Google Scholar]
- Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins: Structure. Function, and Bioinformatics. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Gallagher TM. Murine coronavirus membrane fusion is blocked by modification of thiols buried within the spike protein. J Virol. 1996;70:4683–4690. doi: 10.1128/jvi.70.7.4683-4690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina A, Hanley TM, Mandel R, Trahey M, Broder CC, Viglianti GA, Ryser HJP. Inhibitors of protein-disulfide isomerase prevent cleavage of disulfide bonds in receptor-bound glycoprotein 120 and prevent HIV-1 entry *. J Biol Chem. 2002;277:50579–50588. doi: 10.1074/jbc.M204547200. [DOI] [PubMed] [Google Scholar]
- Garten M, Prevost C, Cadart C, Gautier R, Bousset L, Melki R, Bassereau P, Vanni S. Methyl-branched lipids promote the membrane adsorption of alpha-synuclein by enhancing shallow lipid-packing defects. Phys Chem Chem Phys. 2015;17:15589–15597. doi: 10.1039/C5CP00244C. [DOI] [PubMed] [Google Scholar]
- Gautier R, Bacle A, Tiberti ML, Fuchs PF, Vanni S, Antonny B. PackMem: a versatile tool to compute and visualize interfacial packing defects in lipid bilayers. Biophys J. 2018;115:436–444. doi: 10.1016/j.bpj.2018.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Emerson SU. Importance of amino acid 216 in nonstructural protein 2B for replication of hepatitis A virus in cell culture and in vivo. J Med Virol. 2003;71:7–17. doi: 10.1002/jmv.10457. [DOI] [PubMed] [Google Scholar]
- Guardiani C, Leggio L, Scorciapino MA, de Pinto V, Ceccarelli M. A computational study of ion current modulation in hVDAC3 induced by disulfide bonds. Biochimica Et Biophysica Acta BBA Biomembranes. 2016;1858:813–823. doi: 10.1016/j.bbamem.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Guixà-González R, Rodriguez-Espigares I, Ramírez-Anguita JM, Carrió-Gaspar P, Martinez-Seara H, Giorgino T, Selent J. MEMBPLUGIN: studying membrane complexity in VMD. Bioinformatics. 2014;30:1478–1480. doi: 10.1093/bioinformatics/btu037. [DOI] [PubMed] [Google Scholar]
- He J, Melnik LI, Komin A, Wiedman G, Fuselier T, Morris CF, Starr CG, Searson PC, Gallaher WR, Hristova K, Garry RF, Wimley WC. Ebola virus delta peptide is a viroporin. J Virol. 2017;91:e00438–e517. doi: 10.1128/JVI.00438-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner DE. Structural insights into redox-active cysteine residues of the Src family kinases. Redox Biol. 2021;41:101934. doi: 10.1016/j.redox.2021.101934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmüller H, MacKerell AD. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(33–8):27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Iyer BR, Mahalakshmi R. Hydrophobic characteristic is energetically preferred for cysteine in a model membrane protein. Biophys J . 2019;117:25–35. doi: 10.1016/j.bpj.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jae Yoon S, Jae Il S, Jun Seob K, Yoo Soo Y, Yeon-Kyun S, Kyeong Kyu K, Sangho L, Dae-Hyuk K. Disulfide bond as a structural determinant of prion protein membrane insertion. Mol Cells. 2009;27:673–680. doi: 10.1007/s10059-009-0089-9. [DOI] [PubMed] [Google Scholar]
- Jain S, McGinnes LW, Morrison TG. Thiol/disulfide exchange is required for membrane fusion directed by the Newcastle disease virus fusion protein. J Virol. 2007;81:2328–2339. doi: 10.1128/JVI.01940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jecht M, Probst C, Gauss-Muller V. Membrane permeability induced by hepatitis A virus proteins 2B and 2BC and proteolytic processing of HAV 2BC. Virology. 1998;252:218–227. doi: 10.1006/viro.1998.9451. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- Kalafatovic D, Giralt E. Cell-penetrating peptides: design strategies beyond primary structure and amphipathicity. Molecules (basel, Switzerland) 2017;22:1929. doi: 10.3390/molecules22111929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Elson C, Penfield J, Kirui A, Chen A, Zhang L, Wang T. Integrated solid-state NMR and molecular dynamics modeling determines membrane insertion of human β-defensin analog. Communications Biology. 2019;2:402. doi: 10.1038/s42003-019-0653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key T, Sarker M, de Antueno R, Rainey JK, Duncan R. The p10 FAST protein fusion peptide functions as a cystine noose to induce cholesterol-dependent liposome fusion without liposome tubulation. Biochimica Et Biophysica Acta BBA - Biomembranes. 2015;1848:408–416. doi: 10.1016/j.bbamem.2014.10.020. [DOI] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontiadou H, Mark AE, Marrink SJ. Antimicrobial peptides in action. J Am Chem Soc. 2006;128:12156–12161. doi: 10.1021/ja062927q. [DOI] [PubMed] [Google Scholar]
- Lipkin RB, Lazaridis T. Implicit membrane investigation of the stability of antimicrobial peptide β-barrels and arcs. J Membr Biol. 2015;248:469–486. doi: 10.1007/s00232-014-9759-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker JK, Griffiths G. An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins. J Cell Biol. 1999;144:267–279. doi: 10.1083/jcb.144.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal J, Zhu X, Cui Q, Yethiraj A. Sequence-dependent interaction of β-peptides with membranes. J Phys Chem B. 2010;114:13585–13592. doi: 10.1021/jp1070242. [DOI] [PubMed] [Google Scholar]
- Moyer CL, Nemerow GR. Disulfide-bond formation by a single cysteine mutation in adenovirus protein VI impairs capsid release and membrane lysis. Virology. 2012;428:41–47. doi: 10.1016/j.virol.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva JL, Agirre A, Nir S, Carrasco L. Mechanisms of membrane permeabilization by picornavirus 2B viroporin. FEBS Lett. 2003;552:68–73. doi: 10.1016/S0014-5793(03)00852-4. [DOI] [PubMed] [Google Scholar]
- Ouberai MM, Wang J, Swann MJ, Galvagnion C, Guilliams T, Dobson CM, Welland ME. alpha-Synuclein senses lipid packing defects and induces lateral expansion of lipids leading to membrane remodeling. J Biol Chem. 2013;288:20883–20895. doi: 10.1074/jbc.M113.478297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo EF, Vemparala S, Kuroda K. Cationic spacer arm design strategy for control of antimicrobial activity and conformation of amphiphilic methacrylate random copolymers. Biomacromol. 2012;13:1632–1641. doi: 10.1021/bm300342u. [DOI] [PubMed] [Google Scholar]
- Palermo EF, Vemparala S, Kuroda K. Antimicrobial polymers: molecular design as synthetic mimics of host-defense peptides. In: Scholz C, Kressler J, editors. Tailored polymer architectures for pharmaceutical and biomedical applications. Washington DC: American Chemical Society; 2013. pp. 319–330. [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinot M, Vanni S, Pagnotta S, Lacas-Gervais S, Payet LA, Ferreira T, Gautier R, Goud B, Antonny B, Barelli H. Lipid cell biology. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science. 2014;345:693–697. doi: 10.1126/science.1255288. [DOI] [PubMed] [Google Scholar]
- Pokhrel R, Pavadai E, Gerstman BS, Chapagain PP. Membrane pore formation and ion selectivity of the Ebola virus delta peptide. Phys Chem Chem Phys. 2019;21:5578–5585. doi: 10.1039/C8CP07323F. [DOI] [PubMed] [Google Scholar]
- Polley A, Vemparala S. Partitioning of ethanol in multi-component membranes: effects on membrane structure. Chem Phys Lipids. 2013;166:1–11. doi: 10.1016/j.chemphyslip.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Bam M, Luat E, Jui MS, Ganewatta MS, Shokfai T, Nagarkatti M, Decho AW, Tang C. Macromolecular-clustered facial amphiphilic antimicrobials. Nature. Communications. 2018;9:5231. doi: 10.1038/s41467-018-07651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani G, Kuroda K, Vemparala S. Towards designing globular antimicrobial peptide mimics: role of polar functional groups in biomimetic ternary antimicrobial polymers. Soft Matter. 2021;17:2090–2103. doi: 10.1039/D0SM01896A. [DOI] [PubMed] [Google Scholar]
- Read J, Clancy EK, Sarker M, de Antueno R, Langelaan DN, Parmar HB, Shin K, Rainey JK, Duncan R. Reovirus FAST proteins drive pore formation and syncytiogenesis using a novel helix-loop-helix fusion-inducing lipid packing sensor. PLoS Pathog. 2015;11:e1004962. doi: 10.1371/journal.ppat.1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnin MV, Vasquez-Montes V, Nepal B, Ladokhin AS, Lazaridis T. Experimental and computational characterization of oxidized and reduced protegrin pores in lipid bilayers. J Membr Biol. 2020;253:287–298. doi: 10.1007/s00232-020-00124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, Buchner J, Schaller M, Stange EF, Wehkamp J. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature. 2011;469:419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- Sharma H, Nagaraj R. Human β-Defensin 4 with non-native disulfide bridges exhibit antimicrobial activity. PLoS ONE. 2015;10:e0119525. doi: 10.1371/journal.pone.0119525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, So L-Y, Chen F, Liang J, Chow H-Y, Wong K-Y, Wan S, Jiang T, Yu R. Influences of disulfide connectivity on structure and antimicrobial activity of tachyplesin I. J Peptide Sci. 2018;24:e3087. doi: 10.1002/psc.3087. [DOI] [PubMed] [Google Scholar]
- Shin J-I, Shin J-Y, Kim J-S, Yang Y-S, Shin Y-K, Kweon D-H. Deep membrane insertion of prion protein upon reduction of disulfide bond. Biochem Biophys Res Commun. 2008;377:995–1000. doi: 10.1016/j.bbrc.2008.10.095. [DOI] [PubMed] [Google Scholar]
- Shukla A, Dey D, Banerjee K, Nain A, Banerjee M. The C-terminal region of the non-structural protein 2B from Hepatitis A Virus demonstrates lipid-specific viroporin-like activity. Sci Rep. 2015;5:15884. doi: 10.1038/srep15884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikdar S, Banerjee M, Vemparala S. Effect of cholesterol on the membrane partitioning dynamics of hepatitis A virus-2B peptide. Soft Matter. 2021;17:7963–7977. doi: 10.1039/D1SM01019K. [DOI] [PubMed] [Google Scholar]
- Suhail S, Zajac J, Fossum C, Lowater H, McCracken C, Severson N, Laatsch B, Narkiewicz-Jodko A, Johnson B, Liebau J, Bhattacharyya S, Hati S. Role of oxidative stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) infection: a review. Protein J. 2020;39:644–656. doi: 10.1007/s10930-020-09935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni S, Vamparys L, Gautier R, Drin G, Etchebest C, Fuchs PF, Antonny B. Amphipathic lipid packing sensor motifs: probing bilayer defects with hydrophobic residues. Biophys J. 2013;104:575–584. doi: 10.1016/j.bpj.2012.11.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni S, Hirose H, Barelli H, Antonny B, Gautier R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat Commun. 2014;5:4916. doi: 10.1038/ncomms5916. [DOI] [PubMed] [Google Scholar]
- Vanni S, Riccardi L, Palermo G, De Vivo M. Structure and dynamics of the acyl chains in the membrane trafficking and enzymatic processing of lipids. Acc Chem Res. 2019;52:3087–3096. doi: 10.1021/acs.accounts.9b00134. [DOI] [PubMed] [Google Scholar]
- Vemparala S, Saiz L, Eckenhoff RG, Klein ML. Partitioning of anesthetics into a lipid bilayer and their interaction with membrane-bound peptide bundles. Biophys J . 2006;91:2815–2825. doi: 10.1529/biophysj.106.085324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Adrian L, Garriga D, Buxaderas M, Fraga J, Pereira PJ, Macedo-Ribeiro S, Verdaguer N. Structural basis for host membrane remodeling induced by protein 2B of hepatitis A virus. J Virol. 2015;89:3648–3658. doi: 10.1128/JVI.02881-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin M, Ekström M, Garoff H. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 2004;23:54–65. doi: 10.1038/sj.emboj.7600012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley M, Wootten D, Conner MT, Simms J, Kendrick R, Logan RT, Poyner DR, Barwell J. Lifting the lid on GPCRs: the role of extracellular loops. Br J Pharmacol. 2012;165:1688–1703. doi: 10.1111/j.1476-5381.2011.01629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann C, Kumar A, Lang A, Ohlenschläger O. Cysteines and disulfide bonds as structure-forming units: insights from different domains of life and the potential for characterization by NMR. Front Chem. 2020;8:280. doi: 10.3389/fchem.2020.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth KD, Monje-Galvan V, Warburton LM, Klauda JB. Effect of membrane lipid packing on stable binding of the ALPS Peptide. J Chem Theory Comput. 2019;15:1418–1429. doi: 10.1021/acs.jctc.8b00945. [DOI] [PubMed] [Google Scholar]
- Worch R, Dudek A, Krupa J, Szymaniec A, Setny P. Charged N-terminus of influenza fusion peptide facilitates membrane fusion. Int J Mol Sci. 2018;19:578. doi: 10.3390/ijms19020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeasmin R, Brewer A, Fine LR, Zhang L. Molecular dynamics simulations of human beta-defensin type 3 crossing different lipid bilayers. ACS Omega. 2021;6:13926–13939. doi: 10.1021/acsomega.1c01803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Disulfide bonds affect the binding sites of human β defensin type 3 on negatively charged lipid membranes. J Phys Chem B. 2020;124:2088–2100. doi: 10.1021/acs.jpcb.9b10529. [DOI] [PubMed] [Google Scholar]
- Zokarkar A, Connolly Sarah A, Jardetzky Theodore S, Lamb Robert A. Reversible inhibition of fusion activity of a paramyxovirus fusion protein by an engineered disulfide bond in the membrane-proximal external region. J Virol. 2012;86:12397–12401. doi: 10.1128/JVI.02006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.