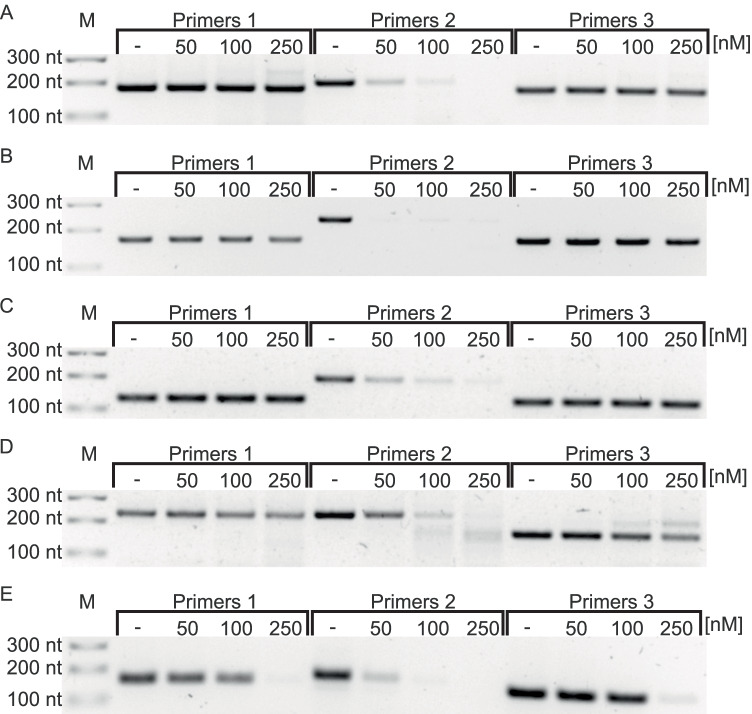
Fig. 3.
SAM riboswitch-containing transcripts are efficiently cleaved by RNase H ex vivo. RNase H cleavage of SAM riboswitch-containing transcripts in the pool of ex vivo transcripts was performed in a gradient of DNA oligomer concentrations (50–250 nM), for each of the analyzed genes: gyrA (A), samT (B), metIC (C), metE (D), and mtnKA (E). A clear disappearance of the signals from products amplified solely by primers 2 (F2/R2) is observed, which indicates specific RNaseH cleavage. The signals derived from the PCR products of primers 1 and 3 are not significantly decreased. M, DNA size marker