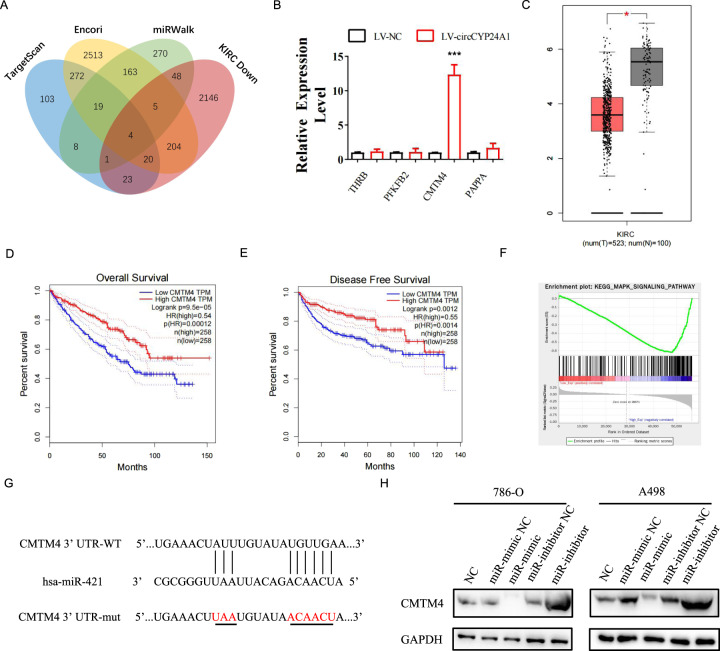
Fig. 4. CMTM4 is the target of miR-421 in RCC.
A The possible targets of miR-421 were predicted by four databases, including TargetScan, Encori, miRWalk, and KIRC Down. KIRC Down represents the downregulated genes in the differential gene expression analysis of TCGA-KIRC database. B Relative expression of THRB, PFKFB2, CMTM4, and PAPPA was measured using qRT-PCR in 786-O cells transfected with LV-NC or LV-circCYP24A1. C CMTM4 TPM expression in the tumor tissues compared with the normal tissues in TCGA-KIRC database. Overall survival (D) and disease-free survival (E) were measured using the Kaplan–Meier analysis. F Gene-set enrichment analysis of the MAPK signaling pathway in patients from TCGA-KIRC with high/low CMTM4 expression. G The predicted binding site for circCYP24A1 and miR-421 is illustrated. Mutant and wild-type CMTM4 constructs for dual-luciferase reporter assays were generated. H CMTM4 expression was detected using Western blotting in 786-O and A498 cells transfected with miR-421 mimic, miR-421 inhibitor, or its normal control. Data are presented as means ± SD. ***p < 0.001.