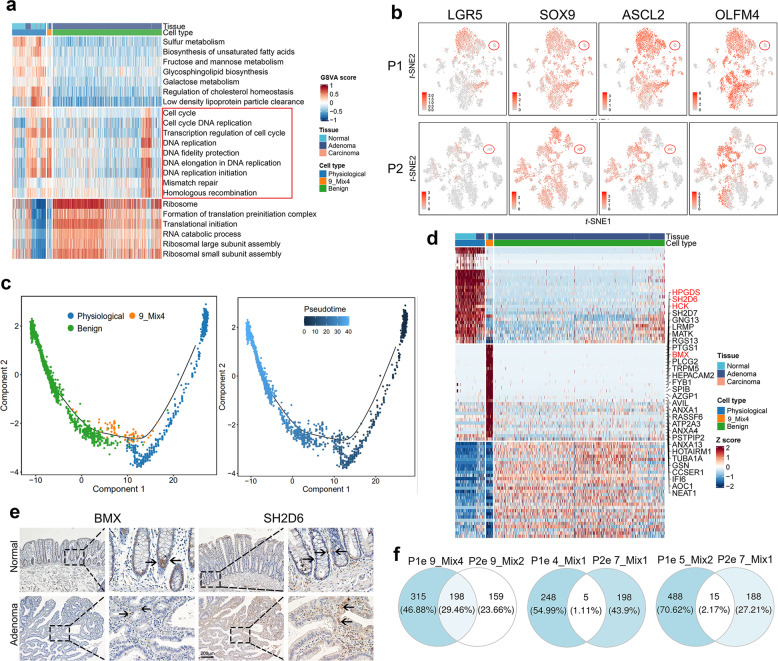
Fig. 4.
Intermediate stages of epithelial cells among the physiological, benign, and malignant stages. a Signature scores for cells from physiological, benign, and mixed clusters (9_Mix4) in P1. Columns represent cells (n = 2420) annotated with cell type and tissue. Colors represent Z scores. b Log-normalized expression levels of key marker genes of intestinal stem and base crypt cells (LGR5, SOX9, ASCL2, OLFM4). Red cycle indicates cells of cluster 9_Mix4 (top, P1, n = 73) and cluster 9_Mix2 (bottom, P2, n = 87). c Single-cell trajectory of 554 physiological epithelial cells, 73 adenoma-precursor cells (cluster 9_Mix4), and 1793 benign epithelial cells. The trajectory was constructed using monocle according to gene expression. Each dot represents a single cell, and colors represent cell types (left) and the pseudotime of trajectory (right). d Heatmap of differentially expressed genes among normal, adenoma precursor (cluster 9_Mix4), and adenoma epithelial cells. Genes related to carcinogenesis are highlighted. e Representative images of BMX and SH2D6 IHC staining of normal tissue (n = 20) and adenoma (n = 20). Black arrows indicate positively stained cells. Scale bar, 200 μm. f Venn diagrams show the overlap of upregulated genes in P1 epithelial mixed clusters (cluster 4, 5, and 9) and P2 epithelial mixed clusters (cluster 7 and 9)