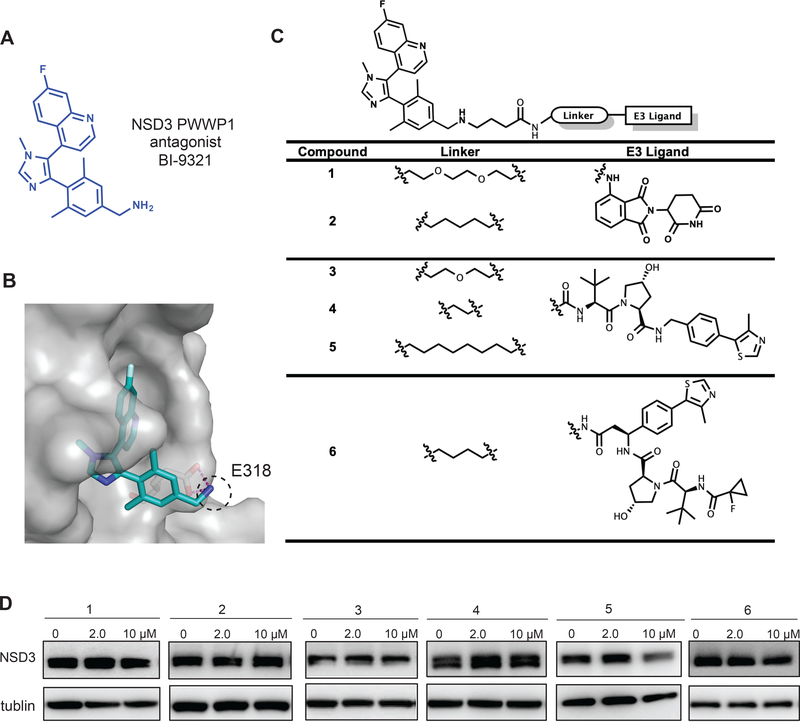
Figure 1. Design and structure–activity relationship results of putative NSD3 PROTACs.
(A) Chemical structure of BI-9321, a selective binder of the NSD3 PWWP1 domain.
(B) Co-crystal structure (PDB code: 6G2O) of NSD3 (gray) in complex with BI-9321 (blue). The primary amino group, highlighted by the dashed black circle, reaches out the binding pocket while making hydrogen bond interactions with E318 of NSD3.
(C) Structures of the putative NSD3 degraders 1–6.
(D) Immunoblotting for NSD3S and GAPDH in 293FT cells after a 48-hour treatment with the indicated concentration of compounds 1–6. Results are representative of at least two independent experiments. See also Figure S1.