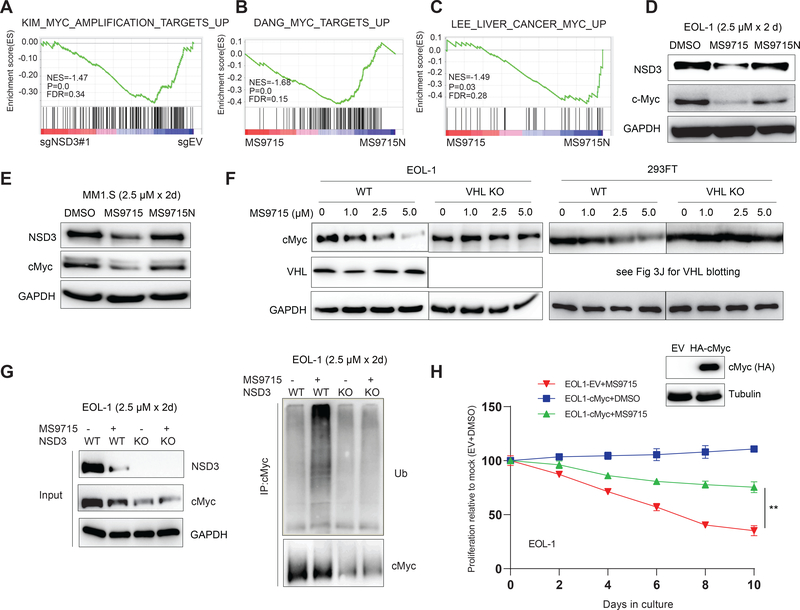
Figure 7. MS9715 represses the cMyc oncogenic node in cancer cells.
(A) GSEA revealing the positive correlation between repression of cMyc-related gene signatures and KO of NSD3 (sg#1), relative to control (sgEV), in EOL-1 cells. The method for calculating P and FDR values in GSEA results is described in STAR Methods section.
(B-C) GSEA revealing the positive correlation between repression of cMyc-related gene signatures and treatment of EOL-1 cells with MS9715, relative to control. See also Figure S5.
(D-E) Immunoblotting for NSD3, cMyc and GAPDH using total cell lysates of EOL-1 (D) and MM1.S (E) cells treated with DMSO or 2.5 μM of MS9715 or MS9715N for 48 hours.
(F) Immunoblot for cMyc and VHL in EOL-1 (left) and 293FT cells (right), either wildtype (WT) or with KO of VHL, after a 48-hour treatment with the indicated concentration of MS9715.
(G) Right: Ubiquitin (Ub) immunoblots (IB) after immunoprecipitation (IP) with cMyc antibodies using total cell lysate of EOL-1 cells, either WT or with KO of NSD3, after a 48-hour treatment with DMSO or 2.5 μM of MS9715. Left: blots for NSD3, cMyc and GAPDH using the input samples of the indicated cells.
(H) Growth of EOL-1 cells, stably expressing either control vector (EV) or HA-tagged cMyc (insert: anti-HA immunoblot), after treatment with DMSO or 2.5 μM of MS9715 for the indicated duration. Y-axis shows the average ± SD of three independent experiments after normalization to DMSO-treated controls. **, P <0.01 by two-sided Student’s t-test.