Summary
Cytosine base editors (CBEs) can install a predefined stop codon at the target site, representing a more predictable and neater method for creating genetic knockouts without altering the genome size. Due to the enhanced predictability of the editing outcomes, it is also more efficient to obtain homozygous mutants in the first generation. With the recent advancement of CBEs on improved editing activity, purify and specificity in plants and animals, base editing has become a more appealing technology for generating knockouts. However, there is a lack of design tools that can aid the adoption of CBEs for achieving such a purpose, especially in plants. Here, we developed a user‐friendly design tool named CRISPR‐BETS (base editing to stop), which helps with guide RNA (gRNA) design for introducing stop codons in the protein‐coding genes of interest. We demonstrated in rice and tomato that CRISPR‐BETS is easy‐to‐use, and its generated gRNAs are highly specific and efficient for generating stop codons and obtaining homozygous knockout lines. While we tailored the tool for the plant research community, CRISPR‐BETS can also serve non‐plant species.
Keywords: cytosine base editors, protospacer adjacent motif, guide RNA design, stop codons, plants
Introduction
Recent development of CRISPR‐Cas‐based genome‐editing tools has revolutionized genetics, medicine and agriculture. Cas9, Cas12a (formerly Cpf1) and Cas12b are all RNA‐guided sequence‐specific nucleases (SSNs) that are easier to engineer and use than earlier SSNs such as zinc finger nucleases (ZFNs) and TAL effector nucleases (TALENs; Malzahn and Lowder, 2017; Voytas, 2013; Zhang et al., 2019). In eukaryotic cells, repair of DNA double strand breaks (DSBs) is predominantly done by the non‐homologous end joining (NHEJ) pathway, which is error prone and generates small insertions and deletions (indels) at the target sites (Featherstone and Jackson, 1999; Puchta, 2005). Such indels often lead to premature stop codons, facilitating genetic knockout of protein‐coding genes, which represent the most popular use of SSNs in nearly all organisms. This is in part due to the low efficiency of the homology recombination pathway (Puchta, 2005; Scully et al., 2019), which limits the use of homology‐directed repair (HDR) in many studies, especially when genetic knockout of protein‐coding genes is the goal.
The emergence of base editors has gradually been changing the genome‐editing technology landscape in eukaryotic organisms including plants (Anzalone and Koblan, 2020; Gurel et al., 2020; Molla and Yang, 2019; Rees and Liu, 2018). Two major types of base editors have been developed and widely used. The first type includes cytosine base editors (CBEs), which were first reported in 2016 (Komor et al., 2016; Ma et al., 2016; Nishida et al., 2016). The second type includes adenine base editors (ABEs) that were first described in 2017 (Gaudelli et al., 2017). Both CBEs and ABEs were based on the CRISPR‐Cas9 system, utilizing cytidine deaminases and adenine deaminases to confer C‐to‐T and A‐to‐G base transition changes in the editing windows respectively. CBEs, not ABEs, can convert four codons (CGA, GAG, GAA and TGG) into stop codons (TGA, TAG, TAA) (Molla and Yang, 2019). Hence, CBEs can be used for knocking out protein‐coding genes by introducing premature stop codons. Generating such stop codons with base editing is more predictable than with CRISPR nucleases that can generate frame shift mutations by NHEJ which are much harder to predict. With high predictability of the editing outcomes, CBEs are also more likely to generate homozygous knockout mutants in the first generation, which is of great significance. In plants, this translates into an accelerated process of getting homozygous mutants for reverse genetics or crop breeding. The use of CBEs for the introduction of stop codons has been demonstrated in many plant species, including rice (Komatsu et al., 2020; Shimatani et al., 2017), wheat (Zong et al., 2018), tomato (Hunziker et al., 2020) and poplar (Li et al., 2021a).
The predominant CBEs used in plants are in the BE3 configuration where a cytidine deaminase and an uracil glycosylase inhibitor (UGI) are fused to a Cas9D10A nickase (Komor et al., 2016). The editing window is generally ~4–10 nucleotides within the ~20 nucleotide protospacer sequence and it can be shifted towards either to the end of the protospacer or narrowed or broadened, depending on the cytidine deaminases and protein fusion configurations in CBEs (Molla and Yang, 2019). For example, different cytidine deaminases have been used in plants such as rABOBEC1 (Li et al., 2017; Lu and Zhu, 2017; Zong et al., 2017), hAID (Ren et al., 2018), PmCDA1 (Shimatani et al., 2017; Tang et al., 2018b), human APOBEC3A (A3A) (Cheng et al., 2021; Zong et al., 2018), APOBEC3B (A3B) (Jin et al., 2020) and A3A/Y130F (Li et al., 2021a; Randall et al., 2021; Ren et al., 2021a). Some comparative studies showed certain cytidine deaminases offered higher base‐editing activities than others in plants. For example, PmCDA1, hAID and A3A were more effective than rABOBEC1 in rice (Tang et al., 2018b; Zong et al., 2018). Recently, we compared seven cytidine deaminases in rice in the BE3 configuration and found that A3A/Y130F conferred the highest C‐to‐T editing activity with a broader editing window (Ren et al., 2021a). Consistent with this, we found that BE3‐A3A/Y130F also generated base‐editing frequency in dicot plants, up to 72.7% in tomato (Randall et al., 2021) and 95.5% in poplar (Li et al., 2021a). Such a high editing frequency is pretty much on par with the efficiency of targeted mutagenesis by Cas nuclease‐mediated NHEJ. Hence, it is promising to apply high‐efficiency CBEs such as BE3‐A3A/Y130F for creating gene knockouts.
Since each CBE has its own preferred editing window, it is very important to broaden the editing scope so that more stop codons can be conveniently introduced by the user‐preferred CBE. Towards this end, Cas9 variants with altered or relaxed protospacer adjacent motif (PAM) requirements can be used. For example, CBEs based on Cas9‐NG (recognizing NG PAMs) were previously demonstrated in rice (Endo et al., 2019; Hua et al., 2019; Li et al., 2021b; Ren et al., 2019a; Zeng et al., 2020; Zhong et al., 2019), as well as in tomato and potato (Veillet et al., 2020). The iSpyMacCas9 (recognizing NAAR PAMs) based CBEs were demonstrated in rice (Sretenovic et al., 2021b), so were CBEs derived from SpCas9‐NRRH, SpCas9‐NRCH and SpCas9‐NRTH (Li et al., 2021b). Furthermore, PAM‐unrestricted SpRY was used to confer PAM‐less C‐to‐T base editing in rice (Li et al., 2021b; Ren et al., 2021b; Xu et al., 2021; Zhang et al., 2021a). Base editing windows and PAM requirements will collectively define all the editable cytosines in the protein coding sequence, and only a fraction of cytosine‐containing codons may be converted to stop codons. Such complexity calls for the development of user‐friendly design tools to aid the design and selection of guide RNAs (gRNAs) for the CBE‐based stop codon introduction in gene knockout experiments.
Earlier, researchers have developed a CRISPR‐STOP gRNA library for introducing stop codons by CBEs in human genes (Kuscu et al., 2017). Similarly, another gRNA database called iStop was developed for introducing stop codons in eight eukaryotic species including one plant species, Arabidopsis thaliana (Billon et al., 2017). Base‐editing design tools have been developed to aid general design in base‐editing experiments, such as CRISPR‐BEST (Tong et al., 2019), BE‐Designer (Hwang et al., 2018) and beditor (Dandage et al., 2019). Additional tools have been developed or adapted for calculating base‐editing frequency, such as BE‐Hive (Arbab et al., 2020), DeepBaseEditor (Song et al., 2020), BE‐Analyzer (Hwang et al., 2018), BEAT (Xu and Liu, 2019) and CRISPResso2 (Clement et al., 2019). So far, only one dedicated design tool, named CRISPR‐CBEI (cytosine base editor‐mediated gene inactivation), has been developed for designing gRNAs to introduce stop codons through C‐to‐T base editing (Yu et al., 2020). The web‐based design tool is interactive and provides a good visualization of the results. However, there are multiple limitations of this software. First, it only allows for DNA input in a Fasta format. Second, it requires uploading of the target genome for assessing off‐target effects of designed gRNAs. This feature makes the tool quite cumbersome to use. Third, CRISPR‐CBEI was not experimentally validated. Finally, the website is often not accessible and hence unreliable, which highlights the vulnerability of an online tool. Considering these limitations, in this study, we aimed to develop a user‐friendly gRNA design tool for introducing stop codons in genes of interests. We call this tool CRISPR‐BETS (base editing to stop), which would provide users best bets for coming up with efficient and specific gRNAs for gene knockout applications in plants and other organisms. There are two versions of CRISPR‐BETS, online (web based) and offline (a PC version, compatible with Win, macOS and Linux). While CRISPR‐BETS may be applied to any genome, we wanted to dedicate it more to the plant research community by including the genomes of 91 plant species. More importantly, we sought to experimentally validate the usefulness of CRISPR‐BETS by testing the designed gRNAs in rice and tomato.
Results
CRISPR‐BETS implementation
To introduce stop codons by CBEs in an organism, three key steps are involved, which are target and molecular reagent selection (step 1), gRNA design (step 2) and wet lab experiments (step 3) (Figure 1a). CRISPR‐BETS was designed to fulfil step 2, which is a critical step that will determine the fate of the downstream wet lab experiments. CRISPR‐BETS has a very simple user interface which can be accessed online (http://zhangtaolab.org/software/crisprbets ) or downloaded to a computer. To start off, a user would input a gene of interest in any of the three formats: GenBank, SnapGene or Fasta (Figure 1b). Then, the user needs to select a CBE system which includes key parameters such as PAM requirements and cytidine deaminases. For different PAM requirements, we included SpCas9 (recognizing NGG PAMs) and its variants such as SpCas9‐VQR (recognizing NGA PAMs), SpCas9‐NG (recognizing NG PAMs), SpCas9‐NRRH (recognizing NRRH PAMs), SpCas9‐NRCH (recognizing NRCH PAMs), SpCas9‐NRTH (recognizing NRTH PAMs) and SpRY (recognizing either NRN PAMs or NYN PAMs). For CBE platforms that incorporate different cytidine deaminases, we included commonly used BE3‐rAPOBEC1, PmCDA‐CBE_V02, BE3‐hAID, BE3‐A3A, BE3‐A3A/Y130F and A3A/Y130F‐CBE_V02 (Ren et al., 2021a). Once the user selects ‘Cas9 variants’ and ‘Deaminase’ options, the following three options termed as PAM’, ‘Edit window’ and ‘gRNA length’ will be prepopulated based on these inputs, which however can be modified, as necessary. Finally, the user would select the genome of interest in ‘Select genome for off‐target analysis’ option, which will output potential off‐target sites for each gRNA designed. Conducting in silico off‐target search could be time consuming. To benchmark CRISPR‐BETS on off‐target search speed, we compared it with the other three software including CRISPR‐CBEI (Yu et al., 2020), Cas‐OFFinder (Bae and Park, 2014) and FlashFry (McKenna and Shendure, 2018). Our test showed that CRISPR‐BETS has outstanding performance over these software (Table S1), enabling fast off‐target assessment. After clicking ‘Analysis’, the software will run, and output designed gRNAs visually. By clicking on each gRNA icon, the gRNA sequence and top off‐target sites can be revealed. The user is just one click away from downloading the whole output file if necessary.
Figure 1.
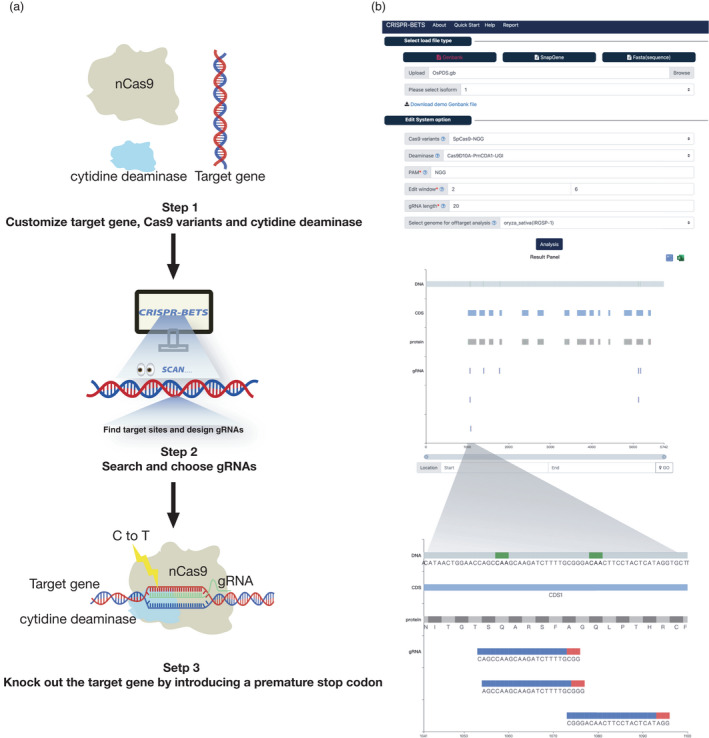
CRISPR‐BETS workflow. (a) Workflow of a base‐editing experiment for creation of premature stop codons. (b) Interface of the CRISPR‐BETS online tool for designing gRNAs tailored for introducing stop codons in protein‐coding genes through C‐to‐T base editing.
Editing Scope of CRISPR‐BETS in plant genomes
To assess the editing scope of CRISPR‐BETS, we conducted in silico analysis of four plant genomes: Arabidopsis thaliana (Arabidopsis), Oryza sativa (rice), Solanum lycopersicum (tomato), and Zea mays (maize). By defining the PAM as NGG (for SpCas9) and the accumulative editing window of 2–12 nt in the protospacer by the available CBEs, we found that 92.00%, 95.30%, 97.31% and 98.21% of genes can be edited to introduce stop codons in tomato, Arabidopsis, rice and maize respectively (Figure 2a). If Cas9 variants with relaxed PAM requirements are used, the percentages of genes that are editable increased at different degrees, reaching nearly 100% in all cases (Figure 2b,c). The earlier a premature stop codon is introduced in the gene, the higher the likelihood of the gene knockout, due to significantly truncated proteins being made and/or more efficient non‐sense‐mediated mRNA decay (NMD) (Brogna and Wen, 2009). Further analysis showed that CRISPR‐BETS can design stop codon‐inducing gRNAs at the first protein‐coding sequence (CDS) or coding exon for about 80%–90% of genes in all four plant species. If the criterion is relaxed to cover before the midpoint/half of ORF (Open Reading Frame), then over 88%–99% of genes can be edited especially when coupled with a relaxed PAM (Figure 2c). Altogether, this analysis of four representative plant genomes suggests wide applicability of CRISPR‐BETS for generating gene knockouts in plants.
Figure 2.
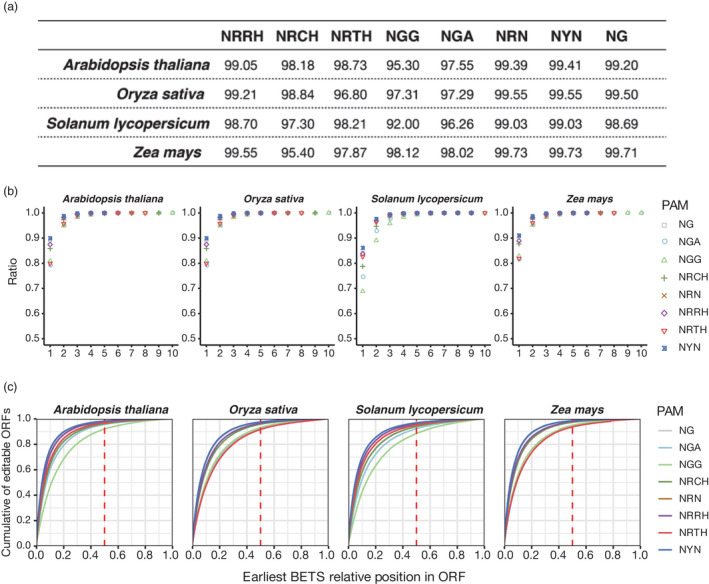
Editing scope applicable by CRISPR‐BETS. (a) The percentages of protein‐coding genes in four plant genomes that premature stop codons can be introduced by C‐to‐T base editing with different PAM‐compatible CBEs as applicable by CRISPR‐BETS. (b) The likelihood (reflected as cumulative ratio of gene in the y‐axis) of inducing premature stop codons in the first CDS (or coding exon), first two CDS, first three CDS, etc. (as indicated in the x‐axis), applicable by CRISPR‐BETS. (c) Relative position of the earliest premature stop codon introduced by CRISPR‐BETS in each of the four plants ORFs with different PAM‐compatible CBEs.
Assessment of CRISPR‐BETS in rice protoplasts
We reasoned that validation of a computational design tool by conducting wet lab experiments is critical in benchmarking the tool and convincing users of the tool usefulness. Furthermore, we may gain useful insights by testing the tool in real experiments as the results may shed light on future improvement of the tool. Since CRISPR‐BETS represents an alternative method for creating genetic knockouts, we were curious about its performance when compared to the conventional NHEJ‐mediated genetic knockout approach. To this end, we decided to assess CRISPR‐BETS with a close comparison to Cas9‐mediated NHEJ mutagenesis. We chose the nCas9‐PmCDA1‐UGI CBE as it has been widely used in plants with high editing efficiency (Li et al., 2021a; Shimatani et al., 2017; Sretenovic et al., 2021a; Tang et al., 2018b; Zhong et al., 2019) and low gRNA‐independent off‐target effects (Ren et al., 2021a). We designed gRNAs with CRISPR‐BETS for three rice genes, OsPDS, OsGW2 and OsGn1a. Two gRNAs (OsPDS‐sgRNA01 and OsPDS‐sgRNA02) for OsPDS, one gRNA (OsGW2‐sgRNA01) for OsGW2 and two gRNAs (OsGn1a‐sgRNA01 and OsGn1a‐sgRNA02) for OsGn1a were identified by CRISPR‐BETS with very few off‐target sites in the rice genome and hence picked for testing in rice (Figure S1a).
Both nCas9‐PmCDA1‐UGI‐based CBE vectors and Cas9‐based nuclease vectors were constructed for each target site. Vectors were tested in rice protoplasts and the editing outcomes were analysed by next‐generation sequencing (NGS) of PCR amplicons. The analysis showed that Cas9 nuclease produced ~10% to 50% NHEJ indel mutations across these five different target sites. Impressively, the nCas9‐PmCDA1‐UGI system generated comparable editing efficiencies at four out of the five target sites (Figure 3a). The base‐editing frequency was lower than the NHEJ mutation frequency at the OsGW2‐sgRNA01 site (Figure 3a). We further analysed the base‐editing outcomes and found that most C‐to‐T base edits indeed resulted in stop codons, and few indels were generated at these target sites (Figure 3b). Analysis of the editing frequencies for all possible editable cytosines in the protospacers showed that the highest editing occurred at the 2–6 nt positions (Figure 3c), consistent with earlier reports with this CBE (Li et al., 2021a; Ren et al., 2021a; Shimatani et al., 2017; Sretenovic et al., 2021a; Tang et al., 2018b; Zhong et al., 2019) and our CRISPR‐BETS design guideline. These data suggest that gRNAs designed by CRISPR‐BETS indeed yield high percentage of stop codon‐containing edits.
Figure 3.
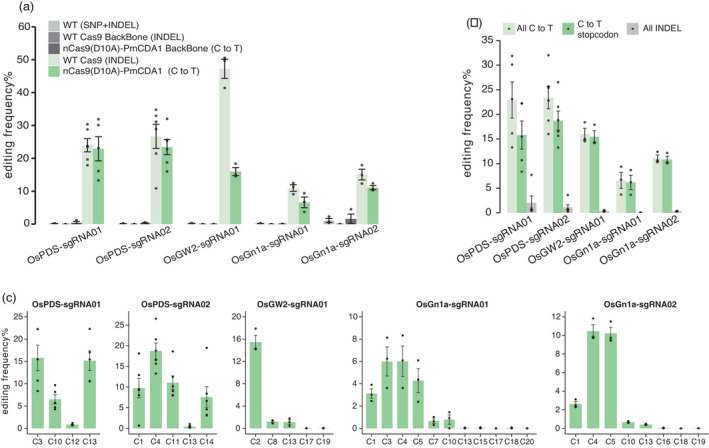
Assessment of CRISPR‐BETS in rice protoplasts. (a) Comparison of Cas9 and nCas9‐PmCDA1‐UGI for genome editing at five different rice loci. Three controls include non‐transformed protoplasts, protoplasts transformed with Cas9 without a gRNA and protoplasts transformed with nCas9‐PmCDA1‐UGI without a gRNA. (b) Comparison efficiencies of C‐to‐T base editing, premature stop codon induction by base editing and indel formation at five target loci. (c) C‐to‐T base‐editing windows by nCas9‐PmCDA1‐UGI at the target sites. The error bars represent standard errors of two to six biological replicates.
Assessment of CRISPR‐BETS in tomato protoplasts
To see whether stop codons can be reliably and frequently generated in a dicot species, we tested CRISPR‐BETS in tomato. We first wanted to edit the Blc gene encoding a beta‐lycopene cyclase. We again chose the nCas9‐PmCDA1‐UGI and the NGG PAM for the base‐editing system. Two CRISPR‐BETS‐designed gRNAs (SlBlc‐NGG‐gR1 and SlBlc‐NGG‐gR2) were assessed in tomato protoplasts for their C‐to‐T base‐editing performance (Figure S1b). We found that at SlBlc‐NGG‐gR1 ~20% C‐to‐T base‐editing frequency was generated and at SlBlc‐NGG‐gR2 ~30% C‐to‐T base‐editing frequency occurred (Figure 4a). Impressively, nearly all base‐editing events carried the introduced stop codons (Figure 4a). Analysis of the base‐editing window showed the high editing positions of 2–6 nt in the protospacers (Figure 4b) which were consistent with the data in rice.
Figure 4.
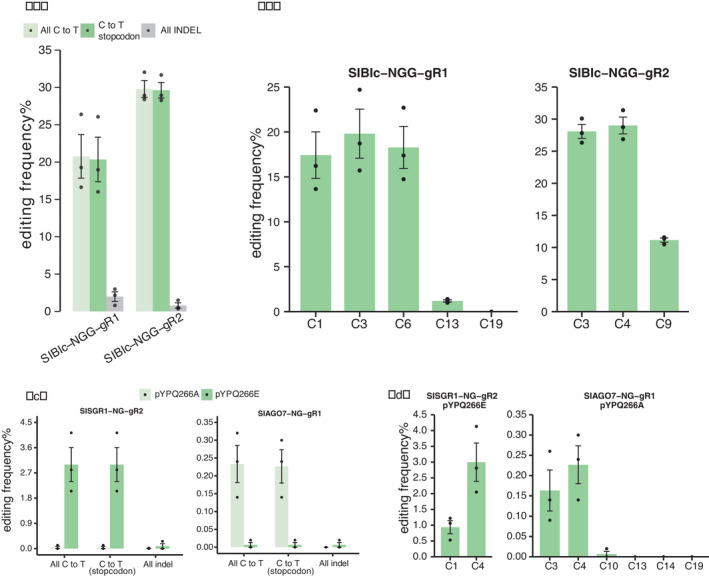
Assessment of CRISPR‐BETS in tomato protoplasts. (a) Comparison of editing frequencies of C‐to‐T base editing, premature stop codon induction and indel formation by nCas9‐PmCDA1‐UGI at two tomato target sites. (b) Editing windows of C‐to‐T base editing by nCas9‐PmCDA1‐UGI at the two target sites. (c) Comparison of C‐to‐T base editing, premature stop codon induction and indel formation by nCas9‐NG‐PmCDA1‐UGI and nSpRY‐PmCDA1‐UGI at two NG target sites. (d) Base editing windows by the two PAM‐relaxed CBEs at the two target sites. The error bars present standard errors of three biological replicates.
Recently, we showed that the use of Cas9‐NG and SpRY could broaden the editing scope of the PmCDA1‐based CBE systems (Ren et al., 2021b; Zhong et al., 2019). We decided to compare both systems at editing of the two relaxed NG PAM sites in tomato protoplasts, one in the STAY‐GREEN 1 (SGR1) gene (Luo et al., 2013) and the other in the AGO7 gene (Husbands et al., 2009). Interestingly, we found Cas9‐NG‐CBE (pYPQ266A) and SpRY‐CBE (pYPQ266E) preferentially edited SlAGO7‐NG‐gR1 and SlSGR1‐NG‐gR2 respectively (Figure 4c). At the SlAGO7‐NG‐gR1 site, Cas9‐NGCBE generated ~0.22% C‐to‐T base‐editing efficiency, while SpRY‐CBE failed at editing this target site. At the SlSGR1‐NG‐gR2 site, SpRY‐CBE generated ~3% base‐editing efficiency, while Cas9‐NG CBE failed at editing this site. It is well known that Cas9‐NG‐ and SpRY‐based base editors have low editing efficiency and high failure rates, which is partly due to their vector self‐editing tendency (Qin et al., 2020; Ren et al., 2021b). However, we found that stop codons were introduced in nearly all the C‐to‐T editing events at both target sites (Figure 4c), which indicated high predictability of CRISPR‐BETS regardless of the Cas9 systems used. As expected, the editing window (Figure 4d) is consistent with other target sites in this study and other studies for nCas9‐PmCDA1‐UGI (Li et al., 2021a; Ren et al., 2021a; Shimatani et al., 2017; Sretenovic et al., 2021a; Tang et al., 2018b; Zhong et al., 2019).
Assessment of CRISPR‐BETS at the OsPDS gene in transgenic rice lines
While the protoplast assays are useful for assessing CRISPR‐BETS, we reasoned that it would be important to further assess the system in stable transgenic plants. Ultimately, one would like to use the design tool to obtain genetic knockout plants. For such assessment, we first carried out the stable transformation of rice with the Cas9 and base‐editing constructs for editing OsPDS with OsPDS‐sgRNA01 and OsPDS‐sgRNA02. Genotyping 18 and 24 T0 lines for Cas9 constructs showed that it generated 100% (18 out of 18 T0 lines) and 54.2% (13 out of 24 T0 lines) indel frequencies at these two target sites respectively (Figure 5a). With nCas9‐PmCDA1‐UGI, 64.0% (16 out of 25 T0 lines) and 40% (10 out of 25 T0 lines) C‐to‐T base‐editing efficiencies were obtained at these two target sites respectively (Figure 5a). Such base‐editing efficiencies, albeit lower than the NHEJ mutagenesis frequencies mediated by Cas9, were sufficient to render base editing as a practical means for gene knockout. We noted that pure C‐to‐T editing frequencies were 40% (10 out of 25 T0 lines) at the OsPDS‐sgRNA01 site and 28% (7 out of 25 T0 lines) at the OsPDS‐sgRNA02 site (Figure 5a). Since biallelic editing of OsPDS would likely generate knockout phenotype, we picked two albino lines each from Cas9‐editing (lines 94‐6‐1 and 94‐7‐1) and nCas9‐PmCDA1‐UGI‐editing (lines 101‐3‐1 and 101‐6‐1) with OsPDS‐sgRNA01 (Figure 5b). Genotyping results showed that the two albino lines resulting from Cas9 editing carried biallelic deletion alleles, and all these alleles presumably destroyed the gene function (Figure 5c). In contrast, those two base‐editing albino lines carried a homozygous TAA stop codon, resulting from simultaneous C‐to‐T base editing of both OsPDS genomic copies (Figure 5c). These data suggest C‐to‐T base editing which, when coupled with the CRISPR‐BETS design, can reliably generate homozygous loss‐of‐function mutants in the first generation.
Figure 5.
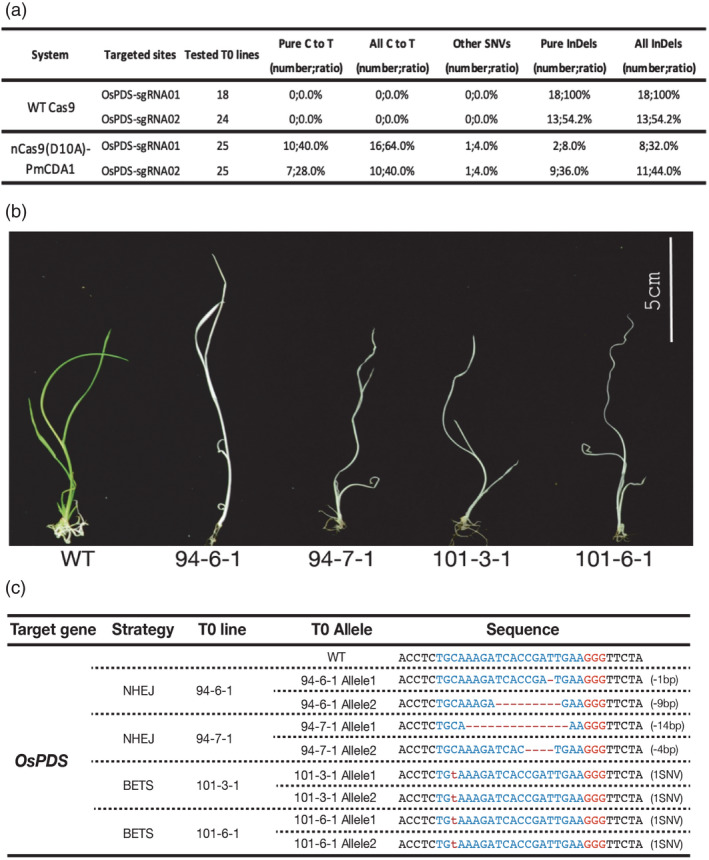
Assessment of CRISPR‐BETS at OsPDS in rice stable lines. (a) A table summarizing genome editing efficiency in rice T0 lines by Cas9 and nCas9‐PmCDA1‐UGI with two sgRNAs targeting OsPDS. (b) Two albino lines derived from Cas9‐mediated mutagenesis, and two albino lines derived from nCas9‐PmCDA1‐UGI‐mediated base editing. (c) Genotypes of the four chosen albino lines reveal homozygosity of base‐edited lines.
Assessment of CRISPR‐BETS at OsGW2 and OsGn1a in transgenic rice lines
We next assessed CRISPR‐BETS at OsGW2 and OsGn1a in stable transgenic rice lines. Cas9 and nCas9‐PmCDA1‐UGI were again closely compared in these cases. At the OsGW2‐sgRNA01 target site, Cas9 generated 93.3% (28 out of 30 T0 lines) indel frequency (Figure 6a). Among the edited lines, 89.3% (25 out of 28) were biallelically edited lines such as 96‐3‐1 and 96‐4‐3 lines (Figure 6b). Base‐editing efficiency by nCas9‐PmCDA1‐UGI at this locus was 16.7% (4 out of 24 T0 lines). Despite relatively low editing efficiency, two out of four base‐editing lines carried a homozygous TAG stop codon due to C‐to‐T base editing. Consistent with the previous reports (Song et al., 2007; Zhou et al., 2019), OsGW2 biallelic mutants by Cas9 and homozygous base‐edited mutants by nCas9‐PmCDA1‐UGI all showed increased seed width and thickness, but not seed length (Figure 6c), which was further supported by quantification of grain length, width and thickness (Figure 6d).
Figure 6.
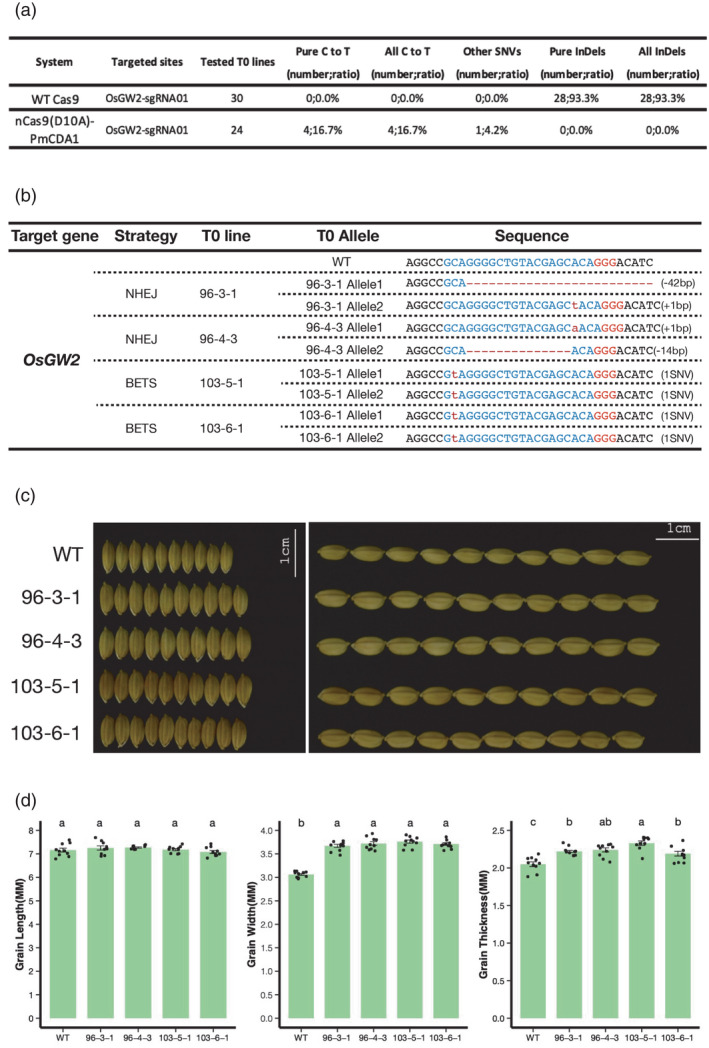
Assessment of CRISPR‐BETS at OsGW2 in rice stable lines. (a) A table summarizing genome editing efficiency in rice T0 lines by Cas9 and nCas9‐PmCDA1‐UGI with one gRNA targeting OsGW2. (b) Genotypes of two biallelic mutants derived from Cas9‐mediated editing and two biallelic homozygous mutants derived from nCas9‐PmCDA‐UGI‐mediated base editing. (c) Phenotypic analysis of seeds from biallelic mutants. (d) Quantification of seed sizes of chosen biallelic mutants. The error bars represent standard errors of 10 biological replicates. Significance was calculated using one‐way ANOVA analysis of variance test, letters denote statistical differences with Tukey’s test (P < 0.05).
At the OsGn1a‐sgRNA01 target site, Cas9 and nCas9‐PmCDA1‐UGI generated 31.6% (6 out of 19 T0 lines) indel frequency and 15.0% (3 out of 30 T0 lines) base‐editing frequency respectively (Figure S2a). At the OsGn1a‐sgRNA02 target site, Cas9 and nCas9‐PmCDA1‐UGI generated comparable editing frequencies: 57.9% (11 out of 19 T0 lines) indel frequency and 56.0% (14 out of 25 T0 lines) respectively (Figure S2a). Genotypic analysis of these edited T0 lines showed that nCas9(D10A)‐PmCDA1, not Cas9, could reliably generate homozygous mutants at the target sites (Figure S2b‐e). Altogether, these data strongly support that CRISPR‐BETS assisted the design of gRNAs for effectively generating homozygous knockout mutants in rice.
Off‐target analysis of CRISPR‐BETS‐designed gRNAs in rice
A potential advantage of CRISPR‐BETS is its built‐in off‐target analysis capability. In an ideal genome‐editing experiment, high editing efficiency should not be compromised by any potential off‐target editing effects. Hence, it is critical to consider off‐target sites when designing and selecting the gRNAs. We have so far tested five sgRNAs for targeted indels and base editing in rice (Table S2). Among them, four are very specific as no more than two computed off‐target sites by CRISPR‐BETS when allowing for up to two nucleotides mismatch to the protospacers (Figure S3a). Analysis of the top off‐target sites of OsPDS‐sgRNA01 and OsGW2‐sgRNA01 in rice protoplasts indeed did not reveal any detectable mutations at these off‐target sites either by Cas9 or by nCas9‐PmCDA1‐UGI (Figure S3b,c). We genotyped the T0 lines corresponding to Cas9 and nCas9‐PmCDA1‐UGI at editing all these five sites. No off‐target mutations were identified for the selected top off‐target sites (Figure S3d). These results suggest that CRISPR‐BETS‐designed gRNAs are highly specific for genome editing.
However, it is well known that Cas9 nuclease may target sequences with one mismatch to the protospacer (Fu et al., 2013, 2014; Tang et al., 2018a). We decided to test a worst‐case scenario by targeting a circle RNA locus in rice with OsCircRNA‐sgRNA01 since there are many off‐target sites that have one nucleotide mismatch to the target site (Figure S4a). Analysis in rice protoplasts showed that both Cas9 and nCas9‐PmCDA1‐UGI resulted in wide‐spread off‐target mutagenesis at these off‐target sites, and surprisingly off‐target mutation frequencies in some cases were higher than the on‐target site (Figure S4b). This example, while not directly related to introducing stop codons in a protein‐coding gene, did highlight the importance of designing highly specific gRNAs to avoid potential off‐target effects. CRISPR‐BETS can facilitate this important design process.
Discussion
In this study, we developed a gRNA design program named CRISPR‐BETS to facilitate the generation of genetic knockouts in plants through C‐to‐T base editing. The data obtained in rice and tomato protoplasts suggest CRISPR‐BETS has a high prediction power as the overall base‐editing efficiency at each target site is nearly equivalent to the efficiency of generating stop codons. We further confirmed such observations in rice stable lines. Although nCas9‐PmCDA1‐UGI generated lower editing efficiency at most of the tested target sites when compared to the targeted mutagenesis by Cas9, its mediated C‐to‐T base editing could effectively generate homozygous mutants in the T0 generation. Such homozygous mutations are much more predictable to abolish the gene function due to the introduction of premature stop codons. Thus, compared to the use of SSNs such as Cas9, Cas12a and Cas12b, C‐to‐T base editing represents a more clean and promising approach for knocking out protein‐coding genes. While nCas9‐PmCDA1‐UGI was used here, we note many CBEs that are likely to confer higher C‐to‐T base‐editing frequency have been recently demonstrated in plants such as A3A (Zong et al., 2018) and A3A‐Y130F (Randall et al., 2021; Ren et al., 2021a). All these CBEs are compatible with CRISPR‐BETS.
Certain CBEs such as BE3 (based on rAPOBEC1) can generate genome‐wide off‐target effects, likely due to cytidine deaminases’ nonspecific binding to DNA (Jin et al., 2019). Genome‐wide off‐target editing is a major concern for clinical applications in humans. Genome‐edited plants could be heavily regulated as conventional genetically modified plants due to safety concerns. CBE‐generated off‐target mutagenesis is often not significant when compared to the somaclonal variation during the plant tissue culture (Jin et al., 2019, 2020; Randall et al., 2021; Ren et al., 2021a) and such off‐target effects may be further reduced by using highly efficient and specific CBEs such as A3Bctd‐VHM‐BE3 (Jin et al., 2020), A3Bctd‐KKR‐BE3 (Jin et al., 2020), PmCDA1‐CBE_V04 (Ren et al., 2021a) and A3A/Y130F‐CBE_V04 (Ren et al., 2021a). Once such high‐specificity CBEs are in use, a practical consideration to avoid off‐target mutations is to design very specific gRNAs. Our data on editing of a circle RNA locus indicated the importance of this issue. Unlike other existing software such as CRISPR‐CBEI (Yu et al., 2020) and CRISPy‐web (Tong et al., 2019), CRISPR‐BETS provides one‐stop analysis of potential off‐target sites for designed gRNAs. Besides covering 91 plant species, CRISPR‐BETS also includes other non‐plant genomes such as Homo sapiens, Mus musculus, Drosophila melanogaster and Saccharomyces cerevisiae. We hope the comprehensive coverage of the plant genomes will facilitate the wide adoption of CRISPR‐BETS by the plant research community.
A great promise of CRISPR‐Cas technologies is the ease of multiplexed genome editing. Multiplexed plant genome‐editing systems were developed for Cas9 (Lowder et al., 2015; Ma et al., 2015; Xie and Minkenberg, 2015; Xing et al., 2014), Cas12a (Hu et al., 2019; Wang et al., 2017, 2018; Zhang et al., 2021b) and Cas12b (Ming et al., 2020). In most of the cases, the goals were for simultaneous knockout of many protein‐coding genes. However, simultaneous generation of multiple DNA DSBs may lead to chromosomal translations, deletions and inversions at variable frequencies (McCarty et al., 2020). These outcomes could be avoided by using base editors as the Cas9 nickase used in CBEs or ABEs would not effectively generate DNA DSBs. Very recently, we showed improved CBEs, such as PmCDA1‐CBE_V04 and A3A/Y130F_CBE_V04, had minimal indel frequencies, indicating further reduced tendency towards generating DNA DSBs (Ren et al., 2021a). A second problem for using a Cas nuclease for generating multiplexed genetic knockouts is the low predictability of the NHEJ‐editing outcomes. As with our data in rice, most if not all biallelic mutations at the target sites by Cas9 are non‐homozygous. While it is relatively straightforward to obtain homozygous mutants in the next generation for one target gene, it would quickly become a big challenge to achieve homozygous triple or quadruple mutants due to the Mendelian segregation of independent mutations. Such genetic segregation may not even be possible or practical in many plant species such as perennial trees. In contrast, high‐efficiency base editing can be achieved in perennial trees such as poplar (Li et al., 2021a). In this regard, C‐to‐T base editing, boosted by CRISPR‐BETS, would be a more effective and appealing approach for rapid generation of homozygous mutants in a multiplexed editing setting, which paves the way for complete knockout of gene families or pathways, in a neat and clean way.
CRISPR‐BETS is an easy‐to‐use software of outstanding performance, which can be upgraded subsequently. In the future, we hope to extend the application of CRISPR‐BETS by adding the support for multiplexed genome editing, since such strategy is important for crop breeding (Zeng et al., 2017; Zhang et al., 2021b). Since NGS amplicon sequencing represents a robust, fast and cheap way for genotyping plants, we also plan to add a PCR primer design feature to aid NGS analysis (Labun et al., 2019), which would greatly ease the workload of wet lab scientists. In addition, we will provide scores for each gRNA (Concordet and Haeussler, 2018), which would help users select the optimal gRNAs for their experiments.
In conclusion, we developed and demonstrated CRISPR‐BETS for implementing C‐to‐T base editing to generate premature stop codons in target genes in plants. CRISPR‐BETS, coupled with the existing and emerging CBEs of high editing efficiency, will greatly promote the use of CBEs for creating single and multiplexed genetic knockout of coding genes in plants and other organisms.
Methods
Development of CRISPR‐BETS
CRISPR‐BETS is written in JavaScript and HTML language, based on the electron framework. To fulfil most of the usage scenarios, CRISPR‐BETS is implemented with both a web version (http://zhangtaolab.org/software/crisprbets) and a computer version, which are integrated into a user‐friendly graphic user interface (GUI) and compatible with major operating systems (Win, macOS, Linux). All dependencies are embedded in CRISPR‐BETS without installation. CRISPR‐BETS allows GenBank, Snapgene(.dna), Fasta file formats as an input. To make the results interactive and easy to view, the gRNA design results are rendered through eCharts (https://echarts.apache.org/zh/index.html). To make the designed gRNAs work more efficiently and broadly applicable, CRISPR‐BETS supports gRNA off‐target prediction in 95 species from ensemble genomes (ftp://ftp.ensemblgenomes.org/pub/plants/), including most of the commonly used plant genomes. Bowtie (Langmead et al., 2009) is used as the backend of CRISPR‐BETS for evaluating the off‐target effect of gRNAs. Exonerate (Slater and Birney, 2005) is utilized to align the CDS sequence back to the DNA sequence to identify the CDS position in the DNA sequence. Detailed documentation of CRISPR‐BETS can be found in https://crispr‐bets‐online.readthedocs.io/en/latest/. A tutorial video was provided on both YouTube and Bilibili (http://zhangtaolab.org/software/crisprbets#video‐tutorial).
Vector construction
For genome editing in rice, the target sites (Table S3) were generated through the online CRISPR‐BETS software. The corresponding oligos (Table S4) were synthesized, annealed and ligated into pGEL031 (Addgene #137900) and pGEL035 (Addgene #137903; Tang et al., 2018b), for targeted mutagenesis by Cas9 and C‐to‐T base editing by nCas9(D10A)‐PmCDA1‐UGI respectively. The resulting T‐DNA expression vectors were summarized in Table S5. The CBE vector systems used in this study were summarized in Table S6.
For base editing in tomato, the target sites (Table S3) were generated by CRISPR‐BETS. Similarly, paired oligos (Table S4) for each target site were generated, annealed and inserted into pYPQ141B (Addgene #69291; Lowder et al., 2015). The nCas9(D10A)‐NG‐PmCDA1 vector, pYPQ266A (Addgene # 173176), was prepared using NEBuilder® HiFi DNA Assembly Master Mix kit (NEB, catalog #E2621*) from PCR‐amplified fragments of vectors pYPQ266 (Ren et al., 2021a) and a zCas9‐NG fragment. The T‐DNA vectors were generated in three‐way Gateway reactions with one attL1‐attR5 base editor entry clone such as pYPQ266 (Addgene #164713), pYPQ266E (Addgene #161521) or pYPQ266A (Addgene # 173176), one attL5‐attL2 pYPQ141B vector containing one cloned gRNA and the attR1‐attR2 destination vector pCGS710, according to our previously established protocol (Sretenovic et al., 2021b).
Rice protoplast isolation and transformation
The Japonica cultivar Kitaake rice seedlings were grown on 1/2 MS solid medium for 12–14 days in the dark at 28°C. The rice protoplast extraction and transformation methods were done by following our previously published protocols (Lowder et al., 2015; Tang et al., 2017). Briefly, the seedling leaves were cut into 0.5–1.0‐mm strips, and put into the enzyme solution. After 30 min of vacuum infiltration, they were incubated at 70–80 rpm for 8 h at 25°C in the dark. The digestion mixture was filtered using a 40‐μm cell strainer. After washing with W5 washing buffer twice, protoplasts were then examined and counted under a microscope. The final protoplast concentration was adjusted to 2 × 106 per millilitre. For protoplast transformation, 30 μg plasmid DNA in 30 μL (1 μg/μL; prepared by Qiagen Midiprep kit) was added to 200 μL protoplasts by gently mixing with 230 μL 40% PEG transformation buffer. After 30 min of incubation in the dark, the reactions were terminated by adding 900 μL W5 washing buffer. The protoplasts were centrifuged and subsequently transferred to a 12‐well culture plate in the dark for 48 h at 32°C.
Tomato protoplast isolation and transformation
The protocol for tomato protoplast isolation and transformation is similar to a recently published protocol (Randall et al., 2021). Briefly, 7–10‐day‐old M82 tomato cotyledons, grown at 25°C (12 h light/12 h dark), were isolated by cutting the petiole where it meets the leaf and incubated in the enzyme solution at 28°C in the dark overnight. The digested cells were filtered by a 75‐μm cell strainer and washed with W5 buffer for three times. The cells were resuspended by 0.55 m sucrose after centrifugation at 200 g for 30 min. The protoplasts were transferred to new 10‐mL tubes and washed with W5 buffer twice. Twenty μg (1000 ng/μL) plasmid DNA was added into 200 μL protoplasts (5 × 105/mL) re‐suspended in MMG and mixed gently. Then, 220 μL of 40% PEG solution was added to the cells and mixed gently. The entire mixture was incubated for 20 min at room temperature. The reactions were stopped by adding 900 μL W5 buffer. Protoplasts were collected by centrifugation and transferred into 12‐well culture plates. Plates were incubated at 32°C in the dark for 60 h. The protoplasts were collected by centrifugation for DNA extraction.
Analysis of base editing in rice and tomato protoplasts
For rice protoplasts, genomic DNA was then extracted with the CTAB method (Ren et al., 2019b; Zhong et al., 2020). For tomato protoplasts, Phire Plant Direct PCR Master Mix (ThermoFisher, USA) was used for direct PCR amplification. Targeted mutagenesis was quantified by next‐generation sequencing (NGS) of PCR amplicons using barcoded primers. The PCR amplicons were sequenced by using an Illumina HiseqX platform. The data were analysed by CRISPRMatch (You et al., 2018).
Rice stable transformation and analysis
The Japonica cultivar Kitaake was used for Agrobacterium‐mediated rice stable transformation as with our previous study (Lowder et al., 2015). The resulting T0 lines were genotyped by Sanger sequencing according to a protocol previously established (Zhou, 2017).
Accession numbers
Gateway® compatible attL1‐attR5 CBE entry vector pYPQ266A has been deposited to Addgene (# 173176). All other vectors have been previously deposited to Addgene. The Next‐generation sequencing (NGS) data have been deposited to the National Center for Biotechnology information (NCBI) database under Sequence Read Archive (SRA) BioProject ‘PRJNA742877’ and Beijing Institute of Genomics Data Center (http://bigd.big.ac.cn) under BioProject ‘PRJCA005698’.
Conflict of interest
Y.Q. is a consultant for Inari Agriculture and CTC Genomics. The remaining authors declare no competing interests.
Author contributions
T.Z., Y.Z. and Y.Q. conceived and designed the experiments. Y.W. developed the CRISPR‐BETS software. Y.W., Y.H. and G.L. built the website. Y.H. and S.L. made the vectors for rice transformation and conducted rice protoplast transformation and data analysis. Y.H., S.L., Q.F., J.Z. and X.Z. did the rice stable transformation. S.S. made the vectors for tomato transformation. Y.C. conducted tomato protoplast transformation and data analysis. Y. W. performed further data analysis and generated the figures. Y.B. assisted with data analysis. T.Z., Y.Z. and Y.Q. supervised the research and wrote the manuscript. All authors participated in discussion and revision of the manuscript.
Supporting information
Figure S1 Target sites in rice and tomato designed by CRISPR‐BETS.
Figure S2 Assessment of CRISPR‐BETS at OsGn1a in rice stable lines.
Figure S3 Target specificity of CRISPR‐BETS‐designed sgRNAs in rice protoplasts and edited T0 lines.
Figure S4 On and off‐target analysis of C‐to‐T base editing for OsCircRNA‐sgRNA01 in rice protoplasts.
Table S1 Benchmark of the off‐target search performance.
Table S2 Off‐target sites evaluated for the gRNAs in rice.
Table S3 Target sites used in this study.
Table S4 Oligos used in this study.
Table S5 T‐DNA constructs used in this study.
Table S6 CBE systems used in this study.
Acknowledgements
The authors thank Drs. Daniel Voytas and Colby Starker for providing the pCGS710 gateway destination vector. This research was supported by the Sichuan Science and Technology Program (2021YFH0084, 2021JDRC0032, 2021YFYZ0016) to J. Z., X. Z. and Y. Z., the National Natural Science Foundation of China (32072045 and 31960423) to X. Z; The Open Foundation of Jiangsu Key Laboratory of Crop Genetics and Physiology (YCSL202009) to J.Z, Y.Z and T.Z. It is also supported by the National Science Foundation Plant Genome Research Program (award no. IOS‐2029889) and the U.S. Department of Agriculture Biotechnology Risk Assessment Grant Program competitive grant (award no. 2018‐33522‐28789) to Y.Q. S. S. is a Foundation for Food and Agriculture Research Fellow.
Wu, Y. , He, Y. , Sretenovic, S. , Liu, S. , Cheng, Y. , Han, Y. , Liu, G. , Bao, Y. , Fang, Q. , Zheng, X. , Zhou, J. , Qi, Y. , Zhang, Y. and Zhang, T. (2022) CRISPR‐BETS: a base‐editing design tool for generating stop codons. Plant Biotechnol. J., 10.1111/pbi.13732
Contributor Information
Yiping Qi, Email: yiping@umd.edu.
Yong Zhang, Email: zhangyong916@uestc.edu.cn.
Tao Zhang, Email: zhangtao@yzu.edu.cn.
References
- Anzalone, A.V. , Koblan, L.W. and Liu, D.R. (2020) Genome editing with CRISPR‐Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844. [DOI] [PubMed] [Google Scholar]
- Arbab, M. , Shen, M.W. , Mok, B. , Wilson, C. , Matuszek, Z. , Cassa, C.A. and Liu, D.R. (2020) Determinants of base editing outcomes from target library analysis and machine learning. Cell, 182, 463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, S. , Park, J. and Kim, J.S. (2014) Cas‐OFFinder: a fast and versatile algorithm that searches for potential off‐target sites of Cas9 RNA‐guided endonucleases. Bioinformatics, 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon, P. , Bryant, E.E. , Joseph, S.A. , Nambiar, T.S. , Hayward, S.B. , Rothstein, R. and Ciccia, A. (2017) CRISPR‐mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol. Cell, 67, 1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna, S. and Wen, J. (2009) Nonsense‐mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 16, 107–113. [DOI] [PubMed] [Google Scholar]
- Cheng, H. , Hao, M. , Ding, B. , Mei, D. , Wang, W. , Wang, H. , Zhou, R. et al. (2021) Base editing with high efficiency in allotetraploid oilseed rape by A3A‐PBE system. Plant Biotechnol. J. 19, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, K. , Rees, H. , Canver, M.C. , Gehrke, J.M. , Farouni, R. , Hsu, J.Y. , Cole, M.A. et al. (2019) CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet, J.P. and Haeussler, M. (2018) CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandage, R. , Despres, P.C. , Yachie, N. and Landry, C.R. (2019) beditor: a computational workflow for designing libraries of guide RNAs for CRISPR‐mediated base editing. Genetics, 212, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, M. , Mikami, M. , Endo, A. , Kaya, H. , Itoh, T. , Nishimasu, H. , Nureki, O. et al. (2019) Genome editing in plants by engineered CRISPR‐Cas9 recognizing NG PAM. Nat. Plants, 5, 14–17. [DOI] [PubMed] [Google Scholar]
- Featherstone, C. and Jackson, S.P. (1999) DNA double‐strand break repair. Curr. Biol. 9, R759–R761. [DOI] [PubMed] [Google Scholar]
- Fu, Y. , Foden, J.A. , Khayter, C. , Maeder, M.L. , Reyon, D. , Joung, J.K. and Sander, J.D. (2013) High‐frequency off‐target mutagenesis induced by CRISPR‐Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. , Sander, J.D. , Reyon, D. , Cascio, V.M. and Joung, J.K. (2014) Improving CRISPR‐Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli, N.M. , Komor, A.C. , Rees, H.A. , Packer, M.S. , Badran, A.H. , Bryson, D.I. and Liu, D.R. (2017) Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature, 551, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel, F. , Zhang, Y. , Sretenovic, S. and Qi, Y. (2020) CRISPR‐Cas nucleases and base editors for plant genome editing. aBIOTECH, 1, 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X. , Meng, X. , Li, J. , Wang, K. and Yu, H. (2019) Improving the efficiency of the CRISPR‐Cas12a system with tRNA‐crRNA arrays. Crop J. 8, 403–407. [Google Scholar]
- Hua, K. , Tao, X. , Han, P. , Wang, R. and Zhu, J.K. (2019) Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol. Plant, 12, 1003–1014. [DOI] [PubMed] [Google Scholar]
- Hunziker, J. , Nishida, K. , Kondo, A. , Kishimoto, S. , Ariizumi, T. and Ezura, H. (2020) Multiple gene substitution by Target‐AID base‐editing technology in tomato. Sci Rep. 10, 20471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands, A.Y. , Chitwood, D.H. , Plavskin, Y. and Timmermans, M.C. (2009) Signals and prepatterns: new insights into organ polarity in plants. Genes Dev. 23, 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, G.H. , Park, J. , Lim, K. , Kim, S. , Yu, J. , Yu, E. , Kim, S.T. et al. (2018) Web‐based design and analysis tools for CRISPR base editing. BMC Bioinformatics, 19, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, S. , Fei, H. , Zhu, Z. , Luo, Y. , Liu, J. , Gao, S. , Zhang, F. et al. (2020) Rationally Designed APOBEC3B Cytosine Base Editors with Improved Specificity. Mol. Cell, 79, 728–740. [DOI] [PubMed] [Google Scholar]
- Jin, S. , Zong, Y. , Gao, Q. , Zhu, Z. , Wang, Y. , Qin, P. , Liang, C. et al. (2019) Cytosine, but not adenine, base editors induce genome‐wide off‐target mutations in rice. Science, 364, 292–295. [DOI] [PubMed] [Google Scholar]
- Komatsu, A. , Ohtake, M. , Shimatani, Z. and Nishida, K. (2020) Production of herbicide‐sensitive strain to prevent volunteer rice infestation using a CRISPR‐Cas9 cytidine deaminase fusion. Front. Plant Sci. 11, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, A.C. , Kim, Y.B. , Packer, M.S. , Zuris, J.A. and Liu, D.R. (2016) Programmable editing of a target base in genomic DNA without double‐stranded DNA cleavage. Nature, 533, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu, C. , Parlak, M. , Tufan, T. , Yang, J. , Szlachta, K. , Wei, X. , Mammadov, R. et al. (2017) CRISPR‐STOP: gene silencing through base‐editing‐induced nonsense mutations. Nat. Methods, 14, 710–712. [DOI] [PubMed] [Google Scholar]
- Labun, K. , Montague, T.G. , Krause, M. , Torres Cleuren, Y.N. , Tjeldnes, H. and Valen, E. (2019) CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res, 47, W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , Trapnell, C. , Pop, M. and Salzberg, S.L. (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Sretenovic, S. , Eisenstein, E. , Coleman, G. and Qi, Y. (2021a) Highly efficient C‐to‐T and A‐to‐G base editing in a Populus hybrid. Plant Biotechnol. J. 1086–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Sun, Y. , Du, J. , Zhao, Y. and Xia, L. (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant, 10, 526–529. [DOI] [PubMed] [Google Scholar]
- Li, J. , Xu, R. , Qin, R. , Liu, X. , Kong, F. and Wei, P. (2021b) Genome editing mediated by SpCas9 variants with broad non‐canonical PAM compatibility in plants. Mol. Plant, 14, 352–360. [DOI] [PubMed] [Google Scholar]
- Lowder, L.G. , Zhang, D. , Baltes, N.J. , Paul, J.W. , Tang, X. , Zheng, X. , Voytas, D.F. et al. (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. and Zhu, J.K. (2017) Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant, 10, 523–525. [DOI] [PubMed] [Google Scholar]
- Luo, Z. , Zhang, J. , Li, J. , Yang, C. , Wang, T. , Ouyang, B. , Li, H. et al. (2013) A STAY‐GREEN protein SlSGR1 regulates lycopene and beta‐carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 198, 442–452. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al. (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Zhang, J. , Yin, W. , Zhang, Z. , Song, Y. and Chang, X. (2016) Targeted AID‐mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods, 13, 1029–1035. [DOI] [PubMed] [Google Scholar]
- Malzahn, A. , Lowder, L. and Qi, Y. (2017) Plant genome editing with TALEN and CRISPR. Cell Biosci. 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, N.S. , Graham, A.E. , Studena, L. and Ledesma‐Amaro, R. (2020) Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 11, 1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A. and Shendure, J. (2018) FlashFry: a fast and flexible tool for large‐scale CRISPR target design. BMC Biol. 16, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming, M. , Ren, Q. , Pan, C. , He, Y. , Zhang, Y. , Liu, S. , Zhong, Z. et al. (2020) CRISPR‐Cas12b enables efficient plant genome engineering. Nat. Plants, 6, 202–208. [DOI] [PubMed] [Google Scholar]
- Molla, K.A. and Yang, Y. (2019) CRISPR/Cas‐mediated base editing: technical considerations and practical applications. Trends Biotechnol. 37, 1121–1142. [DOI] [PubMed] [Google Scholar]
- Nishida, K. , Arazoe, T. , Yachie, N. , Banno, S. , Kakimoto, M. , Tabata, M. , Mochizuki, M. et al. (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science, 353, aaf8729. [DOI] [PubMed] [Google Scholar]
- Puchta, H. (2005) The repair of double‐strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56, 1–14. [DOI] [PubMed] [Google Scholar]
- Qin, R. , Li, J. , Liu, X. , Xu, R. , Yang, J. and Wei, P. (2020) SpCas9‐NG self‐targets the sgRNA sequence in plant genome editing. Nat. Plants, 6, 197–201. [DOI] [PubMed] [Google Scholar]
- Randall, L.B. , Sretenovic, S. , Wu, Y. , Yin, D. , Zhang, T. , Van Eck, J. and Qi, Y. (2021) Genome‐ and transcriptome‐wide off‐target analyses of an improved cytosine base editor. Plant Physiol. 187, 73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees, H.A. and Liu, D.R. (2018) Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, B. , Liu, L. , Li, S. , Kuang, Y. , Wang, J. , Zhang, D. , Zhou, X. et al. (2019a) Cas9‐NG greatly expands the targeting scope of the genome‐editing toolkit by recognizing NG and other atypical PAMs in rice. Mol. Plant, 12, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Ren, B. , Yan, F. , Kuang, Y. , Li, N. , Zhang, D. , Zhou, X. , Lin, H. et al. (2018) Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9‐guided hyperactive hAID mutant. Mol. Plant, 11, 623–626. [DOI] [PubMed] [Google Scholar]
- Ren, Q. , Sretenovic, S. , Liu, G. , Zhong, Z. , Wang, J. , Huang, L. , Tang, X. et al. (2021a) Improved plant cytosine base editors with high editing activity, purity, and specificity. Plant Biotechnol. J. 19, 2052–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Q. , Sretenovic, S. , Liu, S. , Tang, X. , Huang, L. , He, Y. , Liu, L. et al. (2021b) PAM‐less plant genome editing using a CRISPR‐SpRY toolbox. Nat. Plants, 7, 25–33. [DOI] [PubMed] [Google Scholar]
- Ren, Q. , Zhong, Z. , Wang, Y. , You, Q. , Li, Q. , Yuan, M. , He, Y. et al. (2019b) Bidirectional promoter‐based CRISPR‐Cas9 systems for plant genome editing. Front. Plant Sci. 10, 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully, R. , Panday, A. , Elango, R. and Willis, N.A. (2019) DNA double‐strand break repair‐pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani, Z. , Kashojiya, S. , Takayama, M. , Terada, R. , Arazoe, T. , Ishii, H. , Teramura, H. et al. (2017) Targeted base editing in rice and tomato using a CRISPR‐Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443. [DOI] [PubMed] [Google Scholar]
- Slater, G.S. and Birney, E. (2005) Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics, 6, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M. , Kim, H.K. , Lee, S. , Kim, Y. , Seo, S.Y. , Park, J. , Choi, J.W. et al. (2020) Sequence‐specific prediction of the efficiencies of adenine and cytosine base editors. Nat. Biotechnol. 38, 1037–1043. [DOI] [PubMed] [Google Scholar]
- Song, X.J. , Huang, W. , Shi, M. , Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING‐type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Sretenovic, S. , Pan, C. , Tang, X. , Zhang, Y. and Qi, Y. (2021a) Rapid vector construction and assessment of BE3 and Target‐AID C to T base editing systems in rice protoplasts. Methods Mol. Biol. 2238, 95–113. [DOI] [PubMed] [Google Scholar]
- Sretenovic, S. , Yin, D. , Levav, A. , Selengut, J.D. , Mount, S.M. and Qi, Y. (2021b) Expanding plant genome‐editing scope by an engineered iSpyMacCas9 system that targets A‐rich PAM sequences. Plant Commun. 2, 100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X. , Liu, G. , Zhou, J. , Ren, Q. , You, Q. , Tian, L. , Xin, X. et al. (2018a) A large‐scale whole‐genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 19, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X. , Lowder, L.G. , Zhang, T. , Malzahn, A.A. , Zheng, X. , Voytas, D.F. , Zhong, Z. et al. (2017) A CRISPR‐Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants, 3, 17018. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Ren, Q. , Yang, L. , Bao, Y. , Zhong, Z. , He, Y. , Liu, S. et al. (2018b) Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol. J. 17, 1431–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, Y. , Whitford, C.M. , Robertsen, H.L. , Blin, K. , Jorgensen, T.S. , Klitgaard, A.K. , Gren, T. et al. (2019) Highly efficient DSB‐free base editing for streptomycetes with CRISPR‐BEST. Proc. Natl. Acad. Sci. USA, 116, 20366–20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillet, F. , Perrot, L. , Guyon‐Debast, A. , Kermarrec, M.P. , Chauvin, L. , Chauvin, J.E. , Gallois, J.L. et al. (2020) Expanding the CRISPR toolbox in P. patens using SpCas9‐NG variant and application for gene and base editing in solanaceae crops. Int. J. Mol. Sci. 21, 1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas, D.F. (2013) Plant genome engineering with sequence‐specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Mao, Y. , Lu, Y. , Tao, X. and Zhu, J.K. (2017) Multiplex gene editing in rice using the CRISPR‐Cpf1 system. Mol. Plant, 10, 1011–1013. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Mao, Y. , Lu, Y. , Wang, Z. , Tao, X. and Zhu, J.K. (2018) Multiplex gene editing in rice with simplified CRISPR‐Cpf1 and CRISPR‐Cas9 systems. J. Integr. Plant Biol. 60, 626–631. [DOI] [PubMed] [Google Scholar]
- Xie, K. , Minkenberg, B. and Yang, Y. (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proc. Natl. Acad. Sci. USA, 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Liu, Y. and Han, R. (2019) BEAT: a python program to quantify base editing from sanger sequencing. CRISPR J. 2, 223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Kuang, Y. , Ren, B. , Yan, D. , Yan, F. , Spetz, C. , Sun, W. et al. (2021) SpRY greatly expands the genome editing scope in rice with highly flexible PAM recognition. Genome Biol. 22, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Q. , Zhong, Z. , Ren, Q. , Hassan, F. , Zhang, Y. and Zhang, T. (2018) CRISPRMatch: an automatic calculation and visualization tool for high‐throughput CRISPR genome‐editing data analysis. Int. J. Biol. Sci. 14, 858–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. , Wu, Z. , Chen, X. , Ji, Q. and Tao, S. (2020) CRISPR‐CBEI: a designing and analyzing tool kit for cytosine base editor‐mediated gene inactivation. mSystems, 5, e00350‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, D. , Li, X. , Huang, J. , Li, Y. , Cai, S. , Yu, W. , Li, Y. et al. (2020) Engineered Cas9 variant tools expand targeting scope of genome and base editing in rice. Plant Biotechnol. J. 18, 1348–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, D. , Tian, Z. , Rao, Y. , Dong, G. , Yang, Y. , Huang, L. , Leng, Y. et al. (2017) Rational design of high‐yield and superior‐quality rice. Nat. Plants, 3, 17031. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Wang, Y. , Wang, F. , Zhao, S. , Song, J. , Feng, F. , Zhao, J. et al. (2021a) Expanding base editing scope to near‐PAMless with engineered CRISPR/Cas9 variants in plants. Mol. Plant, 14, 191–194. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Malzahn, A.A. , Sretenovic, S. and Qi, Y. (2019) The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants, 5, 778–794. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Ren, Q. , Tang, X. , Liu, S. , Malzahn, A.A. , Zhou, J. , Wang, J. et al. (2021b) Expanding the scope of plant genome engineering with Cas12a orthologs and highly multiplexable editing systems. Nat. Commun. 12, 1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Z. , Liu, S. , Liu, X. , Liu, B. , Tang, X. , Ren, Q. , Zhou, J. et al. (2020) Intron‐based single transcript unit CRISPR systems for plant genome editing. Rice, 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Z. , Sretenovic, S. , Ren, Q. , Yang, L. , Bao, Y. , Qi, C. , Yuan, M. et al. (2019) Improving plant genome editing with high‐fidelity xCas9 and non‐canonical PAM‐targeting Cas9‐NG. Mol. Plant, 12, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Zhou, J. , Deng, K. , Cheng, Y. , Zhong, Z. , Tian, L.I. , Tang, X.U. , Tang, A. et al. (2017) CRISPR‐Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 8, 1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Xin, X. , He, Y. , Chen, H. , Li, Q. , Tang, X. , Zhong, Z. et al. (2019) Multiplex QTL editing of grain‐related genes improves yield in elite rice varieties. Plant Cell Rep. 38, 475–485. [DOI] [PubMed] [Google Scholar]
- Zong, Y. , Song, Q. , Li, C. , Jin, S. , Zhang, D. , Wang, Y. , Qiu, J.L. et al. (2018) Efficient C‐to‐T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953. [DOI] [PubMed] [Google Scholar]
- Zong, Y. , Wang, Y. , Li, C. , Zhang, R. , Chen, K. , Ran, Y. , Qiu, J.L. et al. (2017) Precise base editing in rice, wheat and maize with a Cas9‐ cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Target sites in rice and tomato designed by CRISPR‐BETS.
Figure S2 Assessment of CRISPR‐BETS at OsGn1a in rice stable lines.
Figure S3 Target specificity of CRISPR‐BETS‐designed sgRNAs in rice protoplasts and edited T0 lines.
Figure S4 On and off‐target analysis of C‐to‐T base editing for OsCircRNA‐sgRNA01 in rice protoplasts.
Table S1 Benchmark of the off‐target search performance.
Table S2 Off‐target sites evaluated for the gRNAs in rice.
Table S3 Target sites used in this study.
Table S4 Oligos used in this study.
Table S5 T‐DNA constructs used in this study.
Table S6 CBE systems used in this study.


