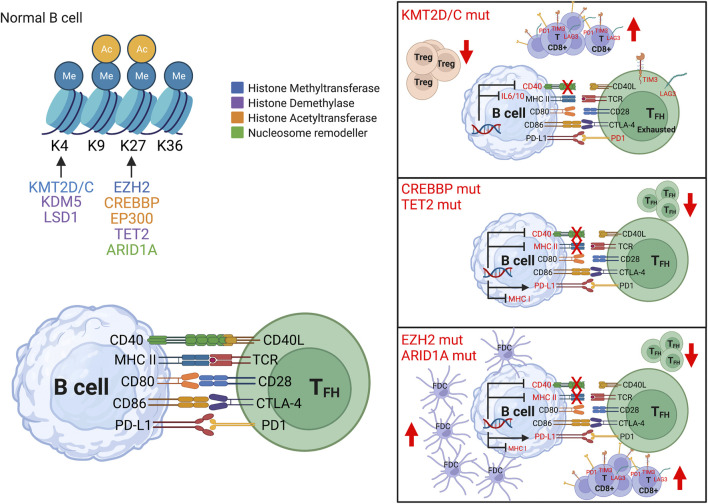
FIGURE 2.
Epigenetic control on immune signaling. In normal B cells, epigenetic mechanisms (DNA methylation, histone modification and chromatin remodeling) control gene expression including immune signaling (left top panel), and in turn modulate immune receptors expression (left bottom panel). In B cell lymphoma, aberrant epigenetic programming can disrupt the immune synapse between B and T cells with concordant reshaping of the immune microenvironment. The most frequent epigenetic alterations are: 1) Mutations in KMT2D/C (right top panel) which suppress CD40, IL10-IL6, NF-kB signaling, with prevalence of exhausted CD8+ T cells and decrease of T regulatory (Treg) cells. 2) Mutations in CREBBP and TET2 (right middle panel) which repress antigen presentation genes (such as MHC class II, its transcription factor CIITA), increase inhibitory molecules (such as PDL1), with lower infiltration of CD4+ T cells. 3) Mutations in EZH2 and ARID1A (right lower panel) which repress CIITA (master regulator of MHC class II), NLRC5 (transactivator of MHC class I) and CD40, with decrease in T follicular helper (TFH) cells and increase in follicular dendritic cells (FDC).