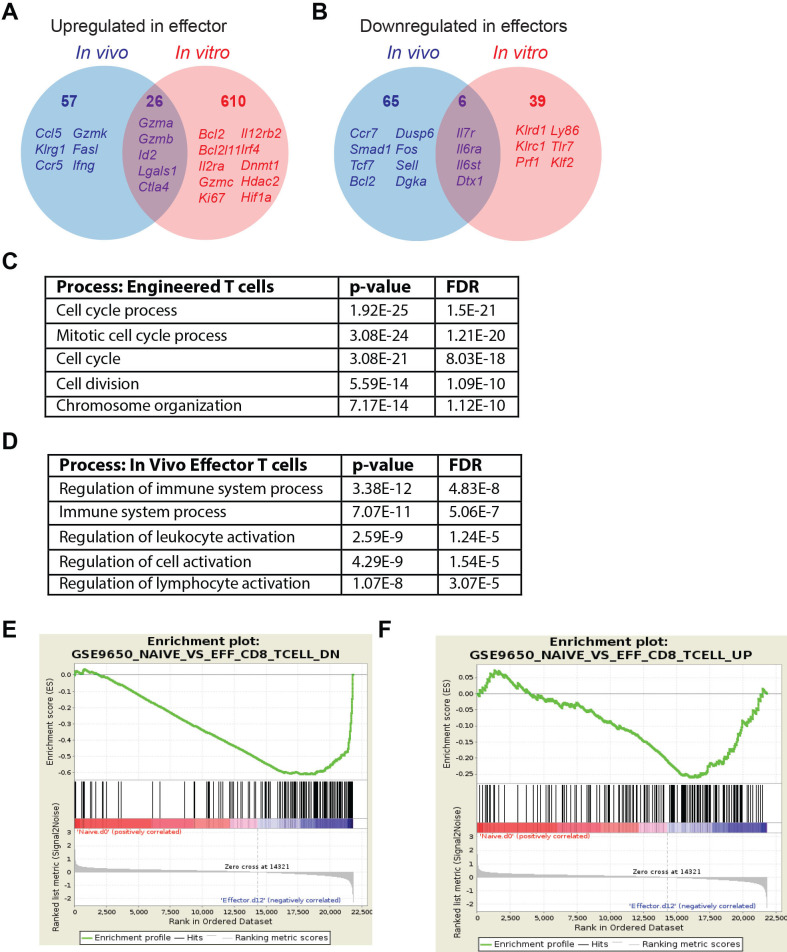
Figure 5.
In vitro-generated effector T cells differ from in vivo-generated effector CD8 T cells. (A) Venn diagrams of genes significantly upregulated in the in vitro-generated engineered CD8 +effector T cells vs day 8 effector CD8 +T cells that arise during acute LCMV infection. All data is normalized to naïve CD8 +T cells. (B) Venn diagrams of genes significantly downregulated in in vitro-generated engineered CD8 +effector T cells vs day 8 effector CD8 +T cells that arise during acute LCMV infection. All data is normalized to naïve CD8 +T cells. (C) Top five biological processes based on DEGs in vivo-derived effector T cells was determined by Gorilla (http://cbl-gorilla.cs.technion.ac.il/). (D) Top five biological processes based on DEGs in vitro-derived engineered T cells was determined by Gorilla (http://cbl-gorilla.cs.technion.ac.il/)/. (E) GSEA of genes downregulated in naïve T cells compared with day 8 in vivo-generated effector T cells vs day 12 in vitro-generated engineered T cells normalized to naïve T cells (GEO:GSE30962). NES, −3.044; p=0; FDR=0. (F) GSEA of genes upregulated in naïve T cells compared with day eight in vivo-generated effector T cells vs day 12 in vitro-generated engineered T cells normalized to naïve T cells (GEO:GSE30962). NES, −1.299; p=0.0578; FDR=0.064. The accession number for microarray data for TCRMsln cells is GSE196435. DEGs, differentially expressed genes; FDR, false discovery rate; GSEA, gene set enrichment analysis; LCMV, lymphocytic choriomeningitis virus; NES, Normalized Enrichment Score.