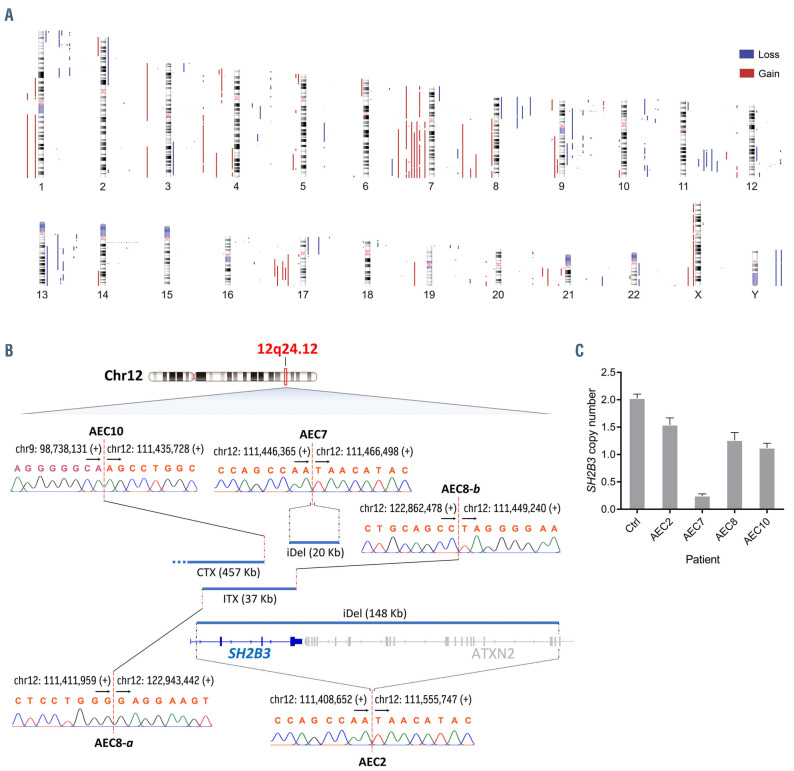
Figure 3.
Landscape of copy number alterations reveals focal SH2B3 inactivation in pcAECyTCL. (A) Human chromosome ideograms showing regions of gain and loss detected through whole-genome sequencing (WGS) in twelve primary cutaneous CD8+ aggressive epidermotropic cytotoxic T-cell lymphoma (pcAECyTCL) genomes. Blue bars to the right of the chromosomes depict regions of loss whereas red bars to the left of the chromosomes depict regions of gain. (B) Deletions at 12q24.12 (blue bars), where SH2B3 resides, were the most focal (<500 Kb) copy number alteration (CNA) events in pcAECyTCL. Inactivation of SH2B3 was mediated by interstitial deletions and unbalanced rearrangements. Breakpoints of structural alterations at 12q24.12 in all affected patients were validated by Sanger sequencing. Genomic coordinates of breakpoints according to reference genome GRCh38. Arrows indicate the direction towards which genomic coordinate numbers increase. Plus (+) and minus (-) signs specify strand polarity. CTX: interchromosomal rearrangement; ITX: intrachromosomal rearrangement; iDel: interstitial deletion. (C) Copy number losses involving SH2B3 in patients with pcAECyTCL were validated by droplet digital polymerase chain reaction. Ctrl: control CD8+ T cells.