Abstract
SARS-CoV-2 is an emerging viral pathogen and a major global public health challenge since December of 2019, with limited effective treatments throughout the pandemic. As part of the innate immune response to viral infection, type I interferons (IFN-I) trigger a signaling cascade that culminates in the activation of hundreds of genes, known as interferon stimulated genes (ISGs), that collectively foster an antiviral state. We report here the identification of a group of type I interferon suppressed genes, including fatty acid synthase (FASN), which are involved in lipid metabolism. Overexpression of FASN or the addition of its downstream product, palmitate, increased viral infection while knockout or knockdown of FASN reduced infection. More importantly, pharmacological inhibitors of FASN effectively blocked infections with a broad range of viruses, including SARS-CoV-2 and its variants of concern. Thus, our studies not only suggest that downregulation of metabolic genes may present an antiviral strategy by type I interferon, but they also introduce the potential for FASN inhibitors to have a therapeutic application in combating emerging infectious diseases such as COVID-19.
Key words: Fatty acid synthase, FASN, IFN-I, SARS-CoV-2, COVID-19, C75, Cerulenin, TVB-3166
Graphical abstract
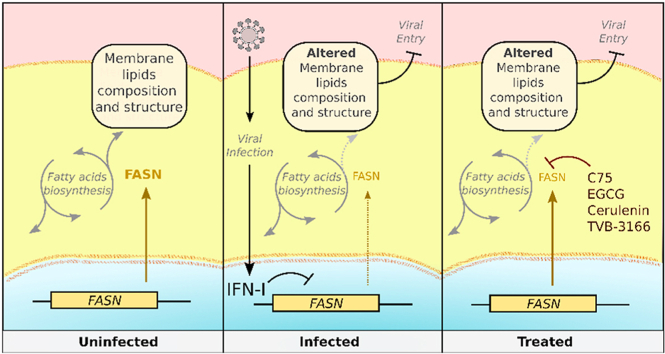
Type I interferon-mediated downregulation of fatty acid synthase (FASN) results in decreased viral infection. Pharmacological inhibitors of FASN could be a novel antiviral therapy against enveloped viruses including SARS-CoV-2.
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is an emerging, positive strand RNA virus from the Coronaviridae family1. As the etiological agent of the coronavirus disease 2019 (COVID-19) pandemic, SARS-CoV-2 has become one of the most challenging public health threats faced in decades, with a major sociological and economic impact2. Despite drastic global efforts to reduce the spread of infection and develop novel treatments and vaccines, the pandemic is to date still uncontrolled, with over 200 million cases and 4.5 million deaths by the end of August, 20213. While the classic presentation of COVID-19 is a respiratory syndrome with a high case-fatality rate, particularly in seniors and those with certain pre-existing health conditions, SARS-CoV-2 infection can also trigger an autoimmune multi-systemic inflammatory disorder, particularly in children4. To combat novel viruses such as SARS-CoV-2 and future emerging viral infections to which we have no specific therapies, it is critical that we develop agents with broad antiviral effects. Investigating innate immune responses, which have evolved to protect the host against multiple types of viral infections, can help us to identify novel strategies to boost or replicate these broad defenses.
The innate immune system provides a critical first line of defense against invading pathogens, including those of viral origin. Utilizing select germ line encoded pattern recognition receptors (PRRs), cells of the innate immune system detect pathogen-associated molecular patterns (PAMPs), which are highly invariable structures of microbial origin, elicits an antiviral gene program known as the host type I interferon (IFN-I) response, characterized by the induction of IFN-I and ISGs. Viral RNA and DNA species generated during replicative cycles serve as PAMPs and are typically detected in the cytosol by nucleic acid sensing PRRs5, 6, 7, 8, 9. Recognition of viral RNA species by the RIG-I like family of RNA sensors, or viral DNA species by the cGAS, DDX41, or IFI16 DNA sensors, triggers the activation of the innate antiviral IFN-I response10, 11, 12. Microbial nucleic acids detected on the cell surface or in endosomal compartments can also activate the host IFN-I response via membrane bound PRRs, such as Toll-like receptors (TLRs) 3, 7, 8, and 9. TLR410,13,14 which detects lipid A and lipopolysaccharide (LPS) derived from the cell walls of gram-negative bacteria, is also known to trigger the IFN-I response15.
A central paradigm of innate antiviral immunity is that IFN-I, namely IFNα and IFNβ, play essential roles in fostering an antiviral state16,17. Operating primarily in a paracrine fashion, these cytokines bind to the IFNα/β receptor (IFNAR) on neighboring cells to instigate a Janus kinase-signal transducer and activator of transcription (JAK–STAT) signaling cascade, which culminates in the up-regulation of nearly 300 ISGs18,19, which directly target viral components or orchestrate cellular processes that lead to the inhibition of virus replication and spread20. Alternatively, certain genes are downregulated by IFNα, β, and γ21 as a mechanism to control viral infections22. Modulation of enzymes involved in the fatty acid synthesis pathway plays a pivotal role in the regulation of infection for many viruses, including human cytomegalovirus (HCMV)23, Kaposi sarcoma-associated herpesvirus (KSHV)24, dengue virus (DENV)25, 26, 27, chikungunya virus28, Rotavirus29, and hepatitis C virus (HCV)30, 31, 32. Utilizing an RNA sequencing (RNAseq) approach, we comparatively evaluated gene expression profiles between wild type (WT) and Ifnar-deficient immune cells after PRR stimulation, and identified fatty acid synthase (Fasn) as an IFN-I suppressed gene that is required for optimal viral infectivity. Our results also reveal that fatty acid synthase (FASN) plays an essential role in mediating viral entry, and that inhibition of FASN by pharmacological compounds represents a novel broad antiviral strategy against a wild range of viruses, including SARS-CoV-2.
2. Materials and methods
2.1. Animals
The experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines from the University of California Los Angeles (CA, USA). Age and sex matched, 6- to 10-week-old mice were used for all experiments. C57Bl/6 and Stat–/– mice were purchased from Jackson laboratories. LysM-Fasn mice were a kind gift from Dr. Semenkovich lab and generated as described33. Tlr3−/−/Cardif–/– double knockout mice were crossed in our laboratory as described previously34.
2.2. Viruses
Dr. Glen Barber (University of Miami, Florida, CA, USA) provided VSV-GFP. MHV68-Luc was provided by Dr. Ren Sun in UCLA (CA, USA). HSV-1, HSV-1 KOS Strain expressing GFP in frame with the ICP0 protein between amino acids 104 and 105 was a kind gift of Dr. William Halford (Southern Illinois University School of Medicine, IL, USA). HSV-1 expressing both GFP and luciferase reporter was a kind gift from Dr. Chunfu Zheng35. The recombinant SARS-CoV-2 (icSARS-CoV-2-mNG) expressing mNeonGreen36 was a kind gift from World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), Department of Microbiology and Immunology, University of Texas Medical Branch through an MTA. SARS-CoV-2 variant of concerns SARS-CoV-2-α (SARS-CoV-2, Isolate hCoV-19/USA/OR-OHSU-PHL00037/2021-B.1.1.7), SARS-CoV-2-β (SARS-CoV-2, Isolate hCoV-19/USA/MD-HP01542/2021B.1.351), SARS-CoV-2-γ (SARS-CoV-2, Isolate hCoV-19/Japan/TY7-503/2021-P.1 or 20J/501Y.V3), and SARS-CoV-2-δ (SARS-CoV-2, Isolate hCoV-19/USA/PHC658/2021-B.1.617.2) were provided by Dr. Vaithilingarajai Arumugaswami who obtained them from the BEI resources.
2.3. Reagents
Luciferase activity was measured using firefly luciferase substrate kit (Promega). Fasn expression plasmids were obtained from Invitrogen, and transfected using Lipofectamine 2000 from Invitrogen according to their protocol. Lipofectamine 2000 was also used for polyI:C transfection (1:1 ratio) in MEFs. Silencer select validated siRNAs were purchased from Ambion and transfected using RNAiMAX transfection reagent from Promega according to their protocol. C75 and cerulenin were from Cayman and EGCG and TVB-3166 were from Sigma. TLR ligands were purchased from Invivogen. All in vitro experiments were repeated at least three times with biological triplicates within each experiment.
2.4. Primary cells and cell lines
A549, Vero, HEK 293 T, and RAW264.7 cells were grown in standard Dulbecco's modified Eagle's medium (DMEM; ThermoFisher) with 10% FBS, 1% penicillin–streptomycin (GIBCO). Bone marrow-derived macrophages (BMDMs) were harvested from 6–8-week C57B/L6 mice (Jackson Labs) and differentiated in DMEM+10%FBS+2%MCSF for 7 days. Media was replaced every other day, and on Day 7 cells were stimulated with ligands. Mouse embryonic-derived fibroblasts (MEF) were derived by skinning the tails of mice and incubating them directly in culture dishes in DMEM with 10% FBS. Cells were scraped and re-plated after 7 days. Vero-E6 and Huh7.5 cell lines were purchased from ATCC, and Hela-ACE2 cells, a kind gift from Dr. Guangxiang Luo from UAB, were cultured in DMEM supplemented with 10% FBS (HyClone), and 1% penicillin–streptomycin (Gibco) at 37 °C in a 5% CO2 humidified atmosphere.
2.5. RNA isolation and RNAseq
5 × 105 BMDMs derived from wild type (C57BL/6) and Ifnar–/– mice were stimulated with Lipid A (100 ng/mL, Sigma) or saline control for 6 h. Cells were harvested in Trizol (Invitrogen) and RNA was isolated by using Qiagen RNeasy Mini Kit (Qiagen) according to manufacturer's protocol. Polyadenylated RNA was purified from 10 μg of total RNA using the Micro-polyA Purist kit (Ambion) according to manufacturer's protocol. Prior to cDNA library construction for RNA-Seq analyses, RNA was quantified and assessed for quality (RNA Integrity Value) using an Agilent 2100 Bioanalyzer (Agilent Technology, Santa Clara, CA, USA). cDNA libraries were constructed as previously described using 100 ng of PolyA + purified RNA as input37,38. All sample sequencing was performed by the Broad Stem Cell Research Center High Throughput Sequencing Core at UCLA on the Illumina HiSeq 2000 with a single end sequencing length of 50 nt. Sequence reads from cDNA libraries were aligned to the mouse genome build NCBI37/mm9 using Tophat39. Alignments were restricted to uniquely mapped reads, with two possible mismatches permitted. RPKM (reads per kilobase per million) values were calculated for mm9 Refseq genes using Seqmonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/). RNAseq data is available at the NCBI GEO database under accession number GSE38892.
2.6. VSV, HSV-1, MHV68, and SARS-CoV-2 viral plaque assay
HEK 293 T cells were infected with VSV-GFP at multiplicity of infection (MOI) of 0.01 for 1 h and the media was replaced with the fresh one. Approximately 150 μL of supernatants were collected at various time points between 8 and 16 h-post infection (hpi) for plaque assay. For HSV-1 and MHV68, 0.25 MOI was used for infection (unless otherwise specified) and supernatants were collected at 24 hpi. Plaque assays were done on Vero cells in 12-well plates at 2 × 105, 5 × 104 and 2 × 104 cells per well for VSV, HSV-1, and MHV68, respectively. Supernatants from infected cells were serially diluted and cells were infected for 1 h. The cells were then covered with growth medium containing low-melting point agarose. Plaques were stained with crystal violet 0.5% (w/v) in 4% PFA (v/v) and were counted after 16 h, 3, and 6 days for VSV, HSV-1, and MHV68, respectively.
Hela cells expressing ACE2 (Hela-ACE-2) were used for SARS-CoV-2 plaque assay. Briefly, media were removed from the confluent mono-layer of Hela-ACE-2 cells followed by infecting cells with the serially diluted virus and incubated for 1 h at 37 °C in a 5% CO2 humidified atmosphere. Cells were then washed with 1 × PBS and then were covered with agarose-overlay. At 72 hpi, the agarose-overlay was removed, cells were washed with 1 × PBS, fixed by 4% PFA for 1 h, and plaques were developed by staining cells with crystal violet containing 1% PFA for 30 min.
2.7. SARS-CoV-2 viral RNA copy number assay
200 μL of the supernatant was harvested from cells infected with SARS-CoV-2 and mixed with equal volume of lysis buffer provided in the high pure viral RNA kit (Invitrogen) and RNA was extracted according to the kit's instructions. The eluted RNA was subjected to RT-qPCR by using the One Step TB Green PrimeScript RT-qPCR Kit II (Takara) and specific primers targeting the SARS-CoV-2 nucleocapsid protein (NP):
Forward, 5′-TAATCAGACAAGGAACTGATTA-3′;
Reverse, 5′-CGAAGGTGTGACTTCCATG-3′.
RT-qPCR cycling conditions were 42 °C for 5 min, 95 °C for 10 s, and 40 cycles of 95 °C for 5 s, followed by 60 °C for 30 s.
All the SARS-CoV-2 based experiments were performed at the UCLA BSL3 facility.
2.8. VSV-G pseudotyped VSV-G luciferase pseudovirus production
VSV-G pseudotyped VSV-G luciferase pseudovirus (VSVΔG-Luc/G) was generated by methods previously described and concentrated by ultracentifugation on 20% sucrose cushion40. The VLPs were resuspended in NTE buffer (100 mmol/L NaCl; 10 mmol/L Tris-HCl, pH 7.5; 1 mmol/L EDTA) and stored in −80 °C. The concentrations used to generate linear range of luciferase signal were determined empirically.
The assembly of hybrid alphavirus SARS-CoV-2 pseudovirus and its variants were described previously41. Briefly, Ha-CoV-2 particles were assembled by cotransfection of HEK 293 T cells in 10 cm dish with 2.5 μg of each of the SARS-CoV-2 structural protein expression vectors (S, N, E, M) and 10 μg of Ha-CoV-2(Luc). Particles were harvested at 48 h post-cotransfection, filtered through a 0.45 μm filter. Ha-CoV-2 particles were used to infect HEK293T-ACE2/TEMPRESS2 cells (a gift from Virongy LLC, Manassas, VA, USA). For C75 inhibition assay, 2.5 × 104 HEK293T-ACE2/TEMPRESS2 cells in each well of 96 well plates were pre-treated with serially diluted C75 for 1 h, cells were infected with Ha-CoV-2(Luc) or its variants of concern for 18 h. Cells were lysed in Luciferase Assay Lysis Buffer (Promega) for luciferase assays using GloMax Discover Microplate Reader (Promega).
2.9. VSVG/βlaM production
A previously described construct encoding Nipah-M1 fused with β-lactamase (βlaM) was used to package inside VSV-G42. HEK 293 T cells were transfected with constructs encoding βlaM and VSV-G or βlaM alone (bald) at 3:1 ratio in 10 cm dishes by polyethylenimine (PEI) transfection reagent. The viral supernatants were collected, clarified, and concentrated by ultracentrifugation at > 75,000 × g on 20% sucrose cushion and the pellet was resuspended in NTE buffer.
2.10. RT-qPCR
Cells were collected in Trizol and RNA was isolated by standard isopropanol precipitation. RNA was quantified and 1 μg of RNA was reversed transcribed using iScript (BioRad) according to the manufacturer's instructions with random hexamer as primers. RT-qPCR analysis was done using the iCycler thermocycler (Bio-Rad). RT-qPCR was conducted in a final volume of 20 μL. Amplification conditions were: 95 °C (3 min), 40 cycles of 95 °C (20 s), 55 °C (30 s), 72 °C (20 s). Expression values were normalized to ribosomal RNA L32 and fold induction was normalized to untreated control.
2.11. Viral RNA detection
For detection of VSV genomic RNA, cells infected with VSV were collected in Trizol and RNA was isolated and reverse transcribed with VSV specific primer N1-5′-GATAGTACCGGAGGATTGACGACTA using Superscript II (Invitrogen) according to manufacturer's protocol. Real time RT-qPCR with Taqman probe with conditions described above. VSV fwd: 5′-GATAGTACCGGAGGATTGACGACTA-3′;
VSV rev: 5′-TCAAACCATCCGAGCCATTC-3′;
VSV probe: 5′-(FAM)-TGCACCGCCACAAGGCAGAGA-(TAMRA)-3′.
For detection of SARS-CoV-2 genomic RNA, the below primers targeting SARS-CoV-2 nucleo capsidprotein (NP) were used:
NP-fwd, 5′-TAATCAGACAAGGAACTGATTA-3′;
NP-rev, 5′-CGAAGGTGTGACTTCCATG-3′.
2.12. MHV68 mouse infections
C57BL/6 mice (6–8 weeks) were purchased from Jackson. Mice were first anesthetized by intraperitoneal (i.p.) injection with 200 mg/kg ketamine, 4 mg/kg xylazine in PBS and shaved. MHV68 (500 pfu) in 200 μL of PBS was administered by i.p. On day three following infection, mice were imaged using the in vivo imaging system (IVIS, Xenogen). Briefly, mice were anesthetized with isoflurane and administered 3 mg d-luciferin/mouse by i.p. Injection prior to imaging. Grayscale photographs and color images of imaged mice were superimposed with LivingImage (Xenogen) and Igor (Wavemetrics) programs, similar to that previously described20. The mice were imaged on dorsal, ventral, right, and left side until the maximal luminescence has passed.
2.13. Generation of FASN–/– A549 cells using CRISPR/Cas9 technology
FASN knockout A549 cell line was obtained using CRISPR/Cas9 system. The target sequences of the two sgRNAs (GCCGGCATGTCCGGGAAGCTGC and CCAGACGCCAGTGTGTGTTCCT) flanked by BsaI restriction site were cloned into px601 plasmid (Addgene) expressing Staphylococcus aureus Cas9 (SaCas9) together with a complete gRNA. Plasmids were then transfected into A549 cells 5 times using lipofectamin 2000 (invitrogene), every three days. The FASN–/– single clone was selected by serial dilution and confirmed by Western blotting.
2.14. Identification of CC50 and IC50 values
Twenty hours prior to the cytotoxicity assay, 8 × 103 Hela-ACE-2, 1.2 × 104 Huh 7.5 and 1.2 × 104 Vero-E6 cells were seeded in 96 Well White/Clear Bottom Plate, TC Surface (Thermo Fisher). Cells were treated with 2-fold serial dilutions (100–1.5 μmol/L) of C75, (160–2.5 μmol/L) of cerulenin, and (160–2.5 μmol/L) of TVB-3166 for 20 h. The cell viability was determined by using Cell Titer-Glo® luminescent cell viability assay (Promega). IC50 and CC50 values were calculated by non-linear regression analysis using GraphPad 5.
2.15. Statistical analysis
The data were analyzed with unpaired student t-test. by Prism software (GraphPad). All of the data are shown as mean + standard deviation (SD) or mean ± standard error of mean (SEM) from three independent experiments. ∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001.
3. Results
3.1. FASN is an IFN-I suppressed gene
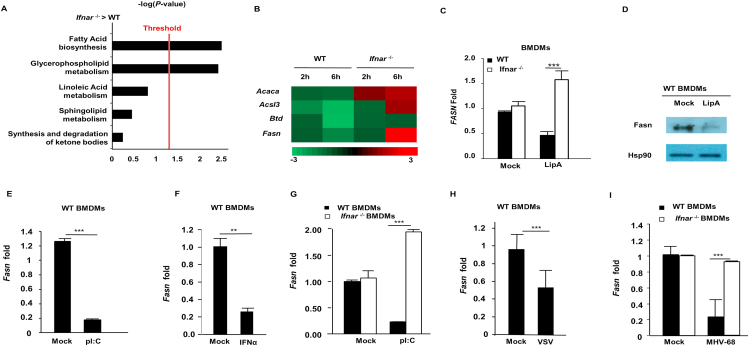
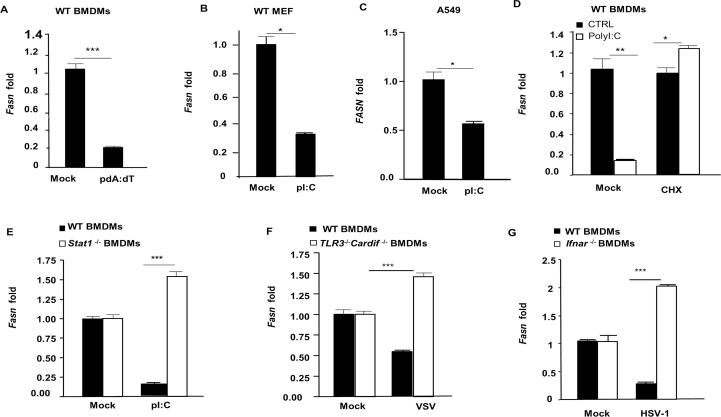
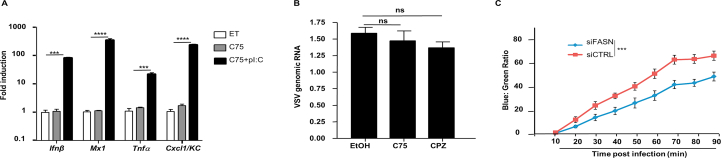
To identify genes that are downregulated by IFN-I during PRR signaling, WT and Ifnar–/– bone marrow-derived macrophages (BMDMs) were stimulated with Lipid A, a TLR4 agonist, and subjected to RNAseq analysis. Ingenuity software profiling of canonical pathways revealed that multiple genes in fatty acid biosynthesis and glycerophospholipid metabolism were down-regulated in WT BMDMs, but upregulated in Ifnar–/– BMDMs (Fig. 1A and B). Accordingly, expression of the gene encoding fatty acid synthase (Fasn) was confirmed by RT-qPCR to be elevated in Ifnar–/– BMDMs and reduced in WT BMDMs, consistent with a decrease in FASN protein levels upon TLR4 stimulation (Fig. 1C and D). Cytosolic PRRs that detect viral nucleic acids to activate the IFN-I response also downregulated Fasn, as BMDMs stimulated with the double stranded (ds) RNA mimetic, polyI:C (pI:C), or dsDNA (poly dA:dT), displayed a marked reduction in Fasn gene expression (Fig. 1E and Supporting Information Fig. S1A). Fasn was also suppressed by IFN-I activating ligands in murine embryonic fibroblasts (MEFs) as well as A549 human lung epithelial cells, suggesting that Fasn downregulation is not restricted to immune cells (Fig. S1B and S1C). TLR4 and cytosolic nucleic acid sensing PRR stimulation elicit IFN-I (Ifnβ/Ifnα) gene transcription, and newly synthesized IFN-I signals via IFNAR to activate ISGs. Downregulation of Fasn requires the synthesis of PRR-induced proteins, as cells treated with the protein translation inhibitor, cycloheximide, failed to reduce Fasn gene expression after PRR stimulation, suggesting that FASN may be suppressed in an IFN-I-dependent fashion (Fig. S1D). Indeed, cells treated with recombinant IFN-α displayed a decrease in Fasn expression (Fig. 1F). Importantly, and consistent with our RNAseq results, PRR ligand-dependent downregulation of Fasn required intact IFNAR and downstream signaling molecules such as STAT1 (Fig. 1G and Fig. S1E). In addition to pure ligands that directly stimulate cytosolic nucleic acid sensing PRRs to induce IFN-I and subsequently suppress FASN, including LipA and pI:C, we examined FASN expression in the context of viral infections. BMDMs infected with vesicular stomatitis virus (VSV), an RNA virus, had a reduction in Fasn gene expression compared to uninfected control cells (Fig. 1H). Downregulation of Fasn during VSV infection required the detection of viral RNA by host PRRs, as cells lacking either MAVS (also known as IPS-1, Cardif, or VISA, a common adaptor molecule that facilitates IFN-I activation downstream of RNA sensing PRRs), or TLR3 (Tlr3−/−/Cardif–/– double knockout cells) were impaired in their ability to suppress Fasn expression (Fig. S1F). Similarly, Fasn was downregulated upon infection with DNA viruses, including murine gamma herpes virus 68 (MHV-68) and herpes simplex virus 1 (HSV-1) in an IFNAR-dependent manner (Fig. 1I and Fig. S1G). Taken together, these findings suggest that Fasn is downregulated by IFN-I during viral infection.
Figure 1.
IFN-I downregulates FASN expression through IFNAR/STAT1 pathways. (A) BMDMs isolated from WT or Ifnar–/– mice were stimulated with LipA (100 ng/mL) for 6 h and RNA samples were isolated and prepared for RNAseq analysis. Using Ingenuity program, canonical pathways which were differentially modulated in Ifnar–/–vs. WT BMDMs were identified. (B) Heat map of fold changes in genes significantly modulated in fatty acid biosynthesis based on the RNAseq data. (C) BMDMs derived from WT and Ifnar–/– mice stimulated with 200 ng/mL LipA and Fasn mRNA levels were assessed by RT-qPCR analysis. (D) WT BMDMs were treated with LipA (200 ng/mL) for 6 h and Fasn protein level was measured by Western blotting. (E) Quantification of Fasn mRNA in WT BMDMs treated with 200 ng/mL pI:C by RT-qPCR. (F) WT BMDMs were treated with IFNα (1000 U/mL) for 6 h and Fasn mRNA was measured by RT-qPCR. (G) Fasn mRNA was quantified in WT and Ifnar–/– BMDMs treated with pI:C (200 ng/mL). (H) The level of Fasn mRNA was measured in WT BMDMs infected with VSV-GFP (MOI = 1) for 8 h. (I) Fasn mRNA was measured in WT and Ifnar–/– BMDMs infected with MHV-68 (MOI = 1) for 8 h. Data presented as mean + SD, n = 3; ∗∗P < 0.01, ∗∗∗P < 0.001.
3.2. FASN supports a broad range of viral infections
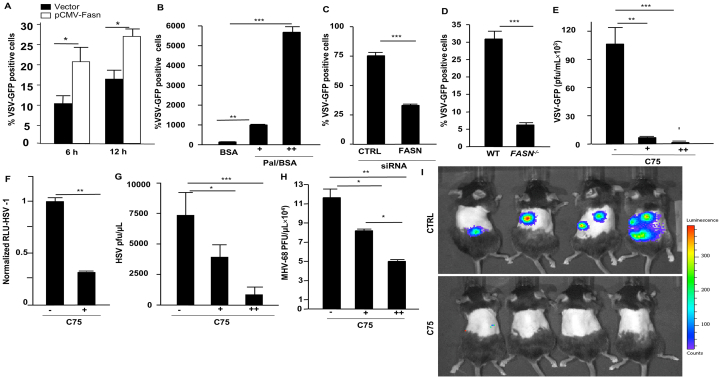
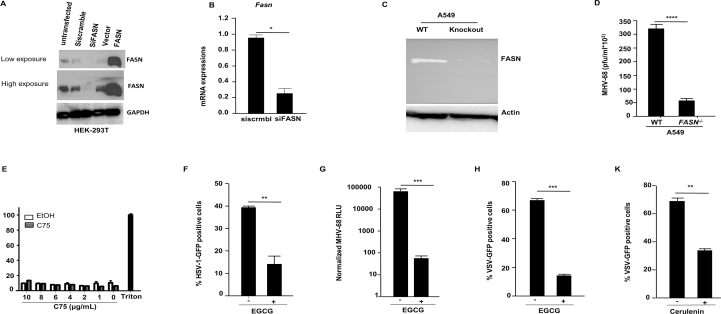
Our data indicate that FASN is suppressed during viral infections in an IFNAR1-dependent manner, suggesting a possibility that it may be involved in modulating viral infection. To determine the role of FASN in antiviral host defense, we ectopically expressed FASN in HEK 293 T cells before infection with different viruses. Overexpression of FASN resulted in a relative increase of viral loads compared to WT infected cells (Fig. 2A and Supporting Information Fig. S2A). HEK 293 T cells supplemented with palmitate, the downstream product of FASN, also displayed an increase in viral infection in a dose-dependent manner (Fig. 2B). In contrast, knockdown of FASN in HEK 293 T cells by small interfering RNA (siRNA) yielded a reduction of viral loads after infection (Fig. 2C, Fig. S2A and S2B). To further validate the functional role of FASN in promoting virus replication, we generated FASN-deficient A549 cells via CRISPR technology (Fig. S2C). In agreement with our siRNA knockdown data, cells lacking FASN failed to support viral infection in comparison to WT control cells (Fig. 2D and Fig. S2D).
Figure 2.
The impact of Fasn expression and activity on viral infection. (A) HEK 293 T cells were transfected with expression plasmids, FASN or control for 36 h prior to VSV infection (MOI = 0.03) and cells were harvested to assess viral infection by FACS. (B) HEK 293 T cells were treated with palmitate/BSA (200 μmol/L) or BSA alone (200 μmol/L) 12 h before VSV infection and cells were harvested at eight hpi for FACS analysis. (C) HEK 293 T cells were transfected with siScramble (control) or siFASN 24 h prior to VSV infection (MOI = 0.03). (D) A549 Fasn–/– and WT cells were infected with VSV-GFP (MOI = 0.03) and cells were subjected to FACS analysis. (E) HEK 293 T cells were treated with C75 (5 and 10 μg/mL) for 10 h before VSV-GFP infections. (F) Human monocyte-derived macrophages were treated with C75 for 12 h (5 μg/mL) prior to HSV-1 (expressing luciferase reporter) infection and viral load was measured by luciferase assay. (G) HEK 293 T cells were treated with C75 (5 and 10 μg/mL) for 12 h prior to HSV-1 infections and viral titer was measured by plaque assay. (H) HEK 293 T cells were treated with C75 (5 and 10 μg/mL) for 12 h before MHV68 infections and viral titer was measured by plaque assay (n = 3). (I) Measuring luciferase activity in mice infected with MHV68luc 8 days post-infection with daily administration of C75 (5 mg/kg, i.p.) or DMEM starting 1 day before infection (n = 5). Data presented as mean + SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
3.3. Pharmacological inhibition of FASN restricts viral infections
C75 (4-methylene-2-octyl-5-oxotetra-hydrofuran-3-carboxylic acid) is a synthetic FASN inhibitor that has been used extensively to study FASN function43. We determined the optimal dose for C75 that would not cause cellular toxicity (Fig. S2E). Cellular treatment with C75 significantly reduced both RNA and DNA viruses in HEK 293 T cells and human monocyte-derived macrophages (hPBMC) compared to untreated cells (Fig. 2E–H). To determine whether inhibition of FASN could inhibit viral infection in vivo, we administered C75 to mice followed by viral infection and full body imaging analysis. Animals treated with C75 harbored lower viral loads after infection compared to untreated animals (Fig. 2I). Epigallocatechin gallate (EGCG), a catechin found in tea leaves (Camellia sinensis)44, and cerulenin, an antifungal antibiotic and irreversible inhibitor of FASN45, have both been reported to function as FASN inhibitors. Similar to C75, cellular treatment with EGCG or cerulenin provided protection against a broad range of enveloped viruses (Fig. S2F–S2K). Together, these results demonstrate that C75 and other pharmacological inhibitors of FASN can effectively inhibit viral infections in vitro and in vivo.
3.4. FASN facilitates viral entry
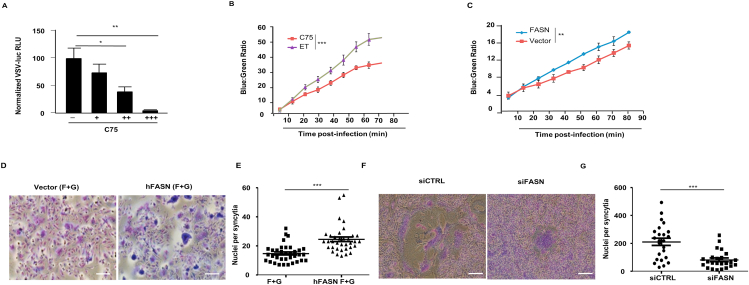
As C75 reduced viral load both in vitro and in vivo, we wanted to determine the mechanism by which FASN operates to promote viral infection. Interestingly, C75 did not suppress viral infection via a mechanism which increases pro-inflammatory or IFN-I cytokines, suggesting that FASN does not function as a steady state negative regulator of the IFN-I response (Supporting Information Fig. S3A). We therefore hypothesized that FASN functions in a manner that does not involve downregulation of innate immune activation pathways, but instead modulates the viral life cycle. Utilizing a pseudo-typed VSV reporter virus (VSVΔG-Luc) that only permits a single round of infection, we infected HEK 293 T cells that were either untreated or treated with the C75 FASN inhibitor. VSVΔG-Luc expression was reduced in cells that were pre-treated with the FASN inhibitor in a dose-dependent manner, indicating that FASN may mediate its pro-viral effects primarily at an early stage in the viral life cycle (Fig. 3A). One of the earliest steps of infection is the attachment of the virus to the host cell membrane. HEK 293 T cells treated with C75, however, showed no significant differences in the number of membrane-attached viral particles when compared to untreated cells, indicating that FASN does not facilitate viral infection at the level of cell membrane attachment (Fig. S3B). We then set out to study the role of FASN on host cell entry, another early step in the viral life cycle. Employing a pseudotyped cytosolic entry reporter VSV-G strain (VSV-G-βlaM), we found that cellular inhibition of FASN via C75 or siRNA inhibited the VSV-G-βlaM cytosolic activity, suggesting that FASN mainly supports viral infection by mediating events involved in viral entry (Fig. 3B and Fig. S3C). Indeed, cells ectopically expressing FASN displayed elevated VSV-G-βlaM cytosolic activity compared to control cells (Fig. 3C). Viral entry requires the fusion of viral particles to the host cell membrane in order to achieve infection. For example, fusion (F) and attachment (G) proteins derived from Nipah virus (NiV) induce pH-independent cell–cell membrane fusion and syncytia formation. Gain of function experiments revealed that FASN enhances membrane fusion and the formation of syncytia between cells (Fig. 3D and E). Alternatively, downregulation of FASN via siRNA results in impaired cell–cell membrane fusion and syncytia formation (Fig. 3F and G). These findings reveal a key role for FASN in early viral infection.
Figure 3.
Mechanisms by which modulation of FASN activity affects viral infection. (A) Infection of C75-Pre-treated HEK 293 T cells with replication deficient pseudo-type VSV-renilla luc. (B) HEK 293 T cells were pretreated with C75 (5 μg/mL) or control (ET) for 12 h or (C) transfected with indicated expression plasmids 36 h prior to VSV-G/βlaM infection for 1.5 h. β-Lactamase activity was measured by determining the rate of cleavage of CCF2 (green) to its cleaved form (blue). (D, E) NiV.F/G syncytia assay, Vero cells were transfected with expression plasmids 24 h prior to NiV F and G transfection and formation of syncytia was determined 36 h after F and G transfection. Scale bar = 100 μm. (F, G) Vero cells were transfected with siRNAs 24 h prior to NiV F and G transfection as well as another round of siRNA transfection. Cells were fixed and syncytia formation was determined 60 h post F and G transfection (n = 3). Scale bar = 100 μm. Data presented as mean + SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.5. FASN inhibitors block SARS-CoV-2 replication
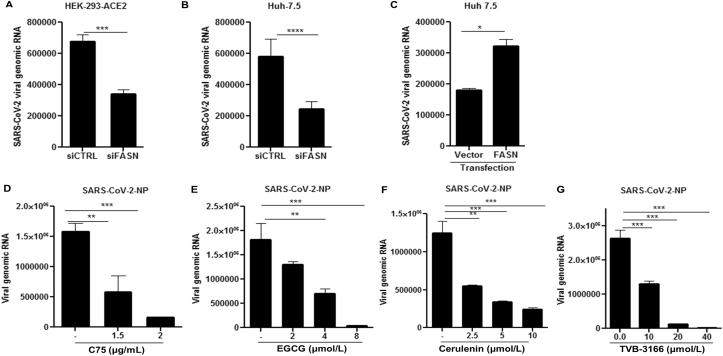
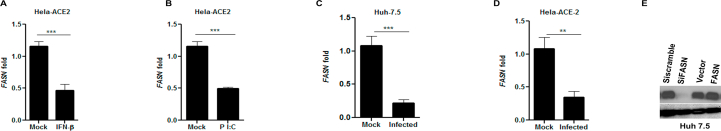
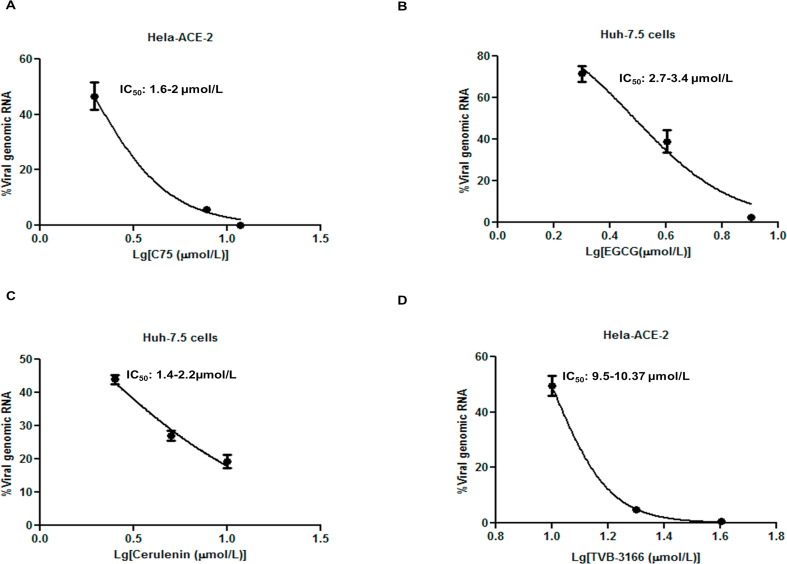
In the setting of the COVID-19 pandemic, we investigated the potential role of FASN in SARS-CoV-2 infection. FASN mRNA levels in Hela-ACE-2 (Hela cells constitutively expressing ACE-2) were reduced after IFNα treatment and pI:C transfection. (Supporting Information Fig. S4A and S4B). The FASN expression was then measured in Hela-ACE-2 and Huh 7.5 cells infected with SARS-CoV-2. We used the recombinant SARS-CoV-2 expressing mNeonGreen protein (icSARS-CoV-2-mNG), as described by Xie et al., 202036. The FASN level was reduced in both cell lines after SARS-CoV-2 infection (Fig. S4C and S4D). To further investigate the role of FASN in this setting, HEK 293 T-ACE2 cells (HEK 293 T cells constitutively expressing ACE2) and Huh 7.5 cells were transfected with siRNA targeting FASN and control siRNA (siScramble) for 36 h followed by SARS-CoV-2 infection. The results demonstrated inhibition of viral infection in FASN-knockdown cells compared to the cells transfected with control siRNA (Fig. 4A and B and Fig. S4E) in both cell lines. Similarly, overexpression of FASN in Huh 7.5 cells increased the viral load compared to cells transfected with vector alone (Fig. 4C and Fig. S4E). We then tested the effect of the FASN inhibitors on SARS-CoV-2 infection in Huh 7.5, Vero-E6, and Hela-ACE-2 cells. Cells were infected with SARS-CoV-2 and 1 h later were treated with C75, EGCG, Cerulenin, or TVB-3166, an analogue of TVB-2640 which is a selective small molecular inhibitor of FASN46.
Figure 4.
SARS-CoV-2 replication can be inhibited by downregulation of FASN. (A) HEK-293 T-ACE2 and (B) Huh 7.5 cells were transfected with siRNA targeting FASN or scramble siRNA (siCTRL) followed by SARS-CoV-2 infection at MOI of 0.01. (C) Transfection of Huh-7.5 cells with a vector expressing FASN or vector alone followed by SARS-CoV-2 infection. (D–G) Huh-7.5 cells were treated with indicated dose of C75, EGCG, cerulenin, and TVB-3166 1 h post SARS-CoV-2 infection. The supernatant was used for titrating the viral RNA copy number and plaque assay. The cell lysate was subjected to RNA extraction to evaluate the viral RNA transcripts by using RT-qPCR (n = 3). All data are means ± SEM; ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
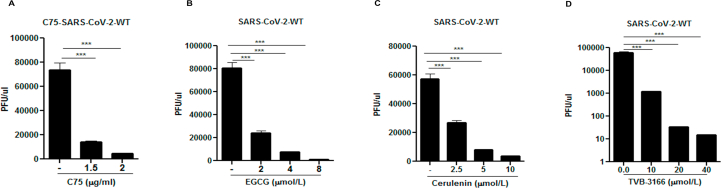
Cells were then harvested and the viral RNA in the cell lysate was quantified by RT-qPCR. Results showed a dose-dependent reduction of SARS-CoV-2 nucleocapsid protein (NP) (Fig. 4D–G). In parallel, infectious viral particles released in the supernatants of the treated cells were measured by standard plaque forming unit assay (PFU) by re-infecting Hela-ACE-2 cells. These results also showed a dose-dependent reduction of viral production in cells treated with the FASN inhibitors (Supporting Information Fig. S5A–S5D).
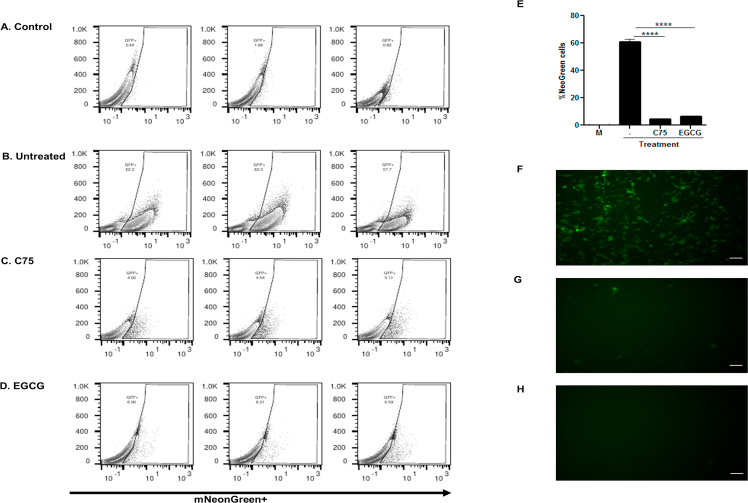
In addition, we used the mNeonGreen recombinant SARS-CoV-2 to infect Vero-E6 and compared the number of GFP positive cells in the presence or absence of FASN inhibitors using flow cytometric analysis and fluorescent microscopy (Supporting Information Fig. S6). Results showed a dramatic reduction in the number of cells infected with the mNeonGreen recombinant SARS-CoV-2 when comparing cells without treatment (61% infected) to 4.5% infection after C75 and 6.6% after ECGC treatments.
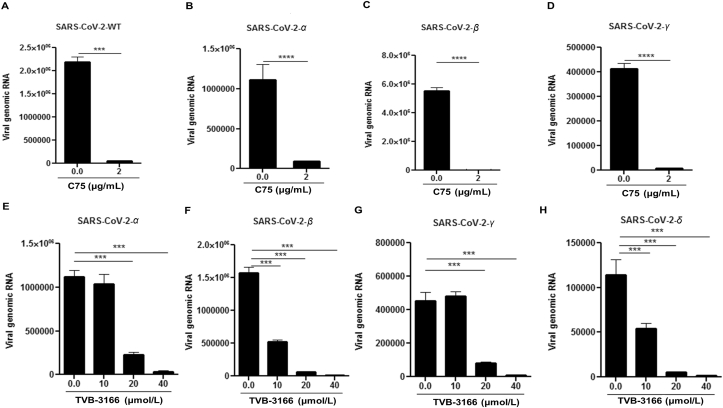
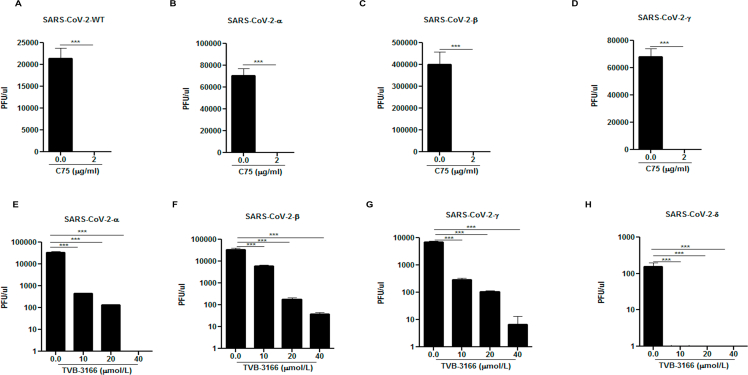
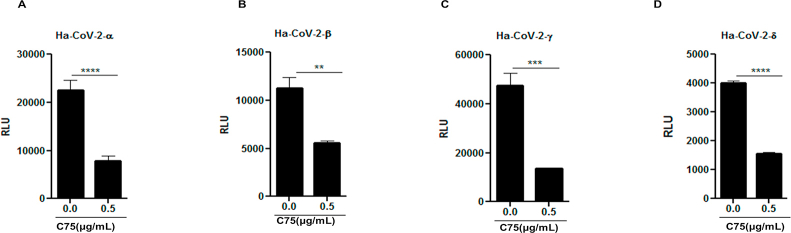
As numerous SARS-CoV-2 variants have appeared since the initial outbreak, we wanted to test whether FASN inhibitors could also effectively suppress infection by these evolved strains, specifically the variants of concern. As shown in Fig. 5, both C75 and TVB-3166 FASN inhibitors suppressed infection by α, β, γ and δ strains at similar efficacies as the WT SARS-CoV-2, based on a viral genomic RNA assay and standard plaque assay (Fig. 5 and Supporting Information Fig. S7). We also obtained similar results with hybrid alphavirus-SARS-CoV-2 pseudo-viruses expressing M, N and E proteins from WT SARS-CoV-2 and S proteins from α, β, γ and δ strains41 which further suggested that FASN inhibitors effectively inhibited the entry of different SARS-CoV-2 variants (Supporting Information Fig. S8). Overall, our studies have provided evidence that FANS inhibitors have potential to be further developed into broad spectrum antiviral agents against multiple viruses, including emerging viruses such as SARS-CoV-2.
Figure 5.
SARS-CoV-2 variants of concern can be inhibited by FASN inhibitors. The Huh 7.5 cells were infected with (A) SARS-CoV-2-WT, (B) SARS-CoV-2-α, (C) SARS-CoV-2-β and (D) SARS-CoV-2-γ at MOI of 0.01 and treated with 2 μg/mL C75 1 h post infection. The Hela-ACE-2 cells were infected with (E) SARS-CoV-2-α, (F) SARS-CoV-2-β, (G) SARS-CoV-2-γ, and (H) SARS-CoV-2-δ at MOI of 0.01 and treated with TVB-3166 at indicated dose 1 h post infection. The cell lysate was harvested for measuring the viral RNA transcripts by using RT-qPCR 24 h post-infection (n = 3). All data are means ± SEM; ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
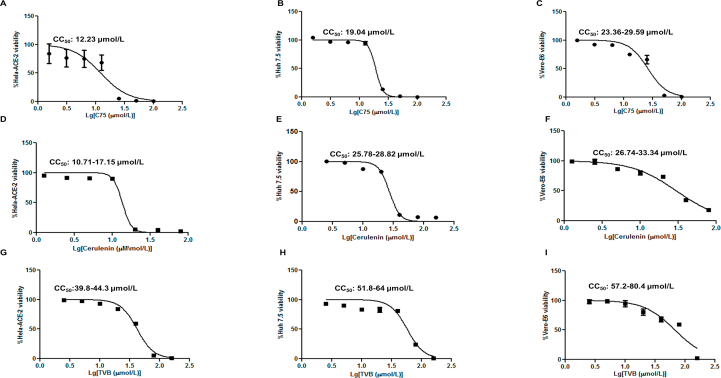
3.6. Identification of CC50 and IC50 values for FASN inhibitors
To determine the potential cytotoxic effect of the compounds used in our study, the 50% cytotoxic concentration (CC50) was evaluated by treating different cell types with 2-fold serial dilutions of C75, cerulenin, and TVB-3166 for 18 h (Supporting Information Fig. S9A–S9I).
We did not detect any apparent cytotoxicity with concentrations of the compounds used to inhibit viral infection in this study. The concentrations used to inhibit 50% of viral infection (IC50) was determined for C75, EGCG, cerulenin, and TVB-3166 by infecting cells with SARS-CoV-2 followed by treating cells with the FASN inhibitors for about 20–24 h (Supporting Information Fig. S10A–S10E). The IC50 for C75 was about 1.6–2 μmol/L, the CC50 was 12–23, 19, and 33.6 μmol/L for Hela-ACE-2, Huh 7.5 and Vero-E6, respectively. The IC50 of EGCG was 2.7–3.4 μmol/L. Cerulenin IC50 was 1.4–2.2 μmol/L, and its CC50 was 10.7, 25.7–28, and 26.7–33 μmol/L for Hela-ACE-2, Huh 7.5 and Vero-E6, respectively. The TVB IC50 was 9.5–10.35 μmol/L and the CC50 was 39.8–44.3, 51.8–64, and 57.2–80.4 μmol/L for Hela-ACE-2, Huh 7.5 and Vero-E6, respectively.
Therefore, the effective dose for viral inhibition for the FASN inhibitors in this study was much smaller than the CC50. However, at higher concentrations there was an apparent toxicity for these inhibitors of FASN (Fig. S9A–S9I).
4. Discussion
Viruses rely on the metabolic network of the host cell, commandeering nucleotide biosynthetic pathways as well as glucose and lipid metabolic pathways to fuel their replication. Some viruses even alter the host's cellular metabolism to favor their own life cycle. For instance, human cytomegalovirus (HCMV) infection upregulates fatty acid biosynthesis and glycolysis, while herpes simplex virus-1 (HSV-1) appears to inhibit glycolytic flux and directs host metabolic pathways towards enhanced pyrimidine synthesis47,48. The efficiency of HCV assembly is mainly determined by the droplet-binding domain of the HCV core protein49. Additionally, recent studies suggest that both Kaposi's sarcoma-associated herpes virus (KSHV) and West Nile virus (WNV) induce lipogenesis50,51 and long term lipid metabolism alteration has also been documented in patients recovered from SARS-CoV-152. We have shown that over-expression of FASN facilitates viral infection, whereas knockout or knockdown of FASN inhibits it. The role of FASN in promoting the replication of many RNA and DNA viruses, including human cytomegalovirus (HCMV)23, Kaposi sarcoma-associated herpesvirus (KSHV)24, cytomegalovirus (CMV)23, dengue virus (DENV)26, chikungunya virus (CHIKV)28, and HIV and HCV30,31 has been previously studied.
While viruses have evolved to alter specific host metabolic pathways, it is unclear whether or how host cells alter their own metabolism to protect themselves from viral infection. Innate antiviral immunity operates via an IFNAR dependent manner, where IFN-I induction triggers the activation of a JAK–STAT pathway resulting in the formation of the STAT1–STAT2–IRF9 transcription factor complex, which binds to interferon stimulatory response elements (ISRE) on ISGs to positively drive their transcription53,54. It is believed that innate antiviral immunity is mainly mediated via IFN-I-dependent induction of ISGs, which collectively foster an antiviral state. We and others have previously identified numerous antiviral genes by screening through hundreds of ISGs19,20,55. In this report, we have demonstrated that Fasn is downregulated in response to not only LPS and pI:C stimulations, but also by different viral infections. Our results have further shown that Fasn is suppressed not only in an IFNAR-dependent manner, but also in a STAT1-dependent manner. How the STAT1 transcription factor functions to suppress Fasn gene transcription is unknown and is currently under investigation. More importantly, our studies suggest that the mechanism by which IFN-I downregulates certain metabolic genes includes commandeering part of the innate immune antiviral program.
Fatty acids are essential molecules in cell membranes, membranous organelles, signaling, and energy storage. FASN catalyzes the synthesis of palmitate (C16:0), a long chain fatty acid that influences the physical properties of the cell by modulating the fluidity of membrane lipid bilayers. FASN deficiency leads to changes in the composition of the cell membrane33. Viral infection requires optimal membrane fluidity, which leads to the clustering of receptors for cell entry56, 57, 58. We have shown that FASN inhibitors suppress viral entry by blocking fusion between viral and cellular membranes. Additionally, downregulation of FASN by siRNA results in decreasing the viral infection rate, though not as pronounced as that of the pharmacological inhibitors of FASN. The difference may be due to limitations in our study for transient transfection in different cell types, or because of possible off target effects of FASN inhibitors. We have also found that FASN inhibitors can suppress cell–cell membrane fusion and syncytia formation driven by viral proteins, impacting viral infection in multiple stages of the viral life cycle. The results of this study highlight the promising antiviral potency of FASN inhibitors, and their potential role in developing novel antiviral therapeutics. Interestingly, certain viruses co-opt host palmitoyltransferases to modify viral proteins to support optimal infection59. These enzymes regulate palmitoylation (aka S-acylation), a post-translational modification yielding the covalent linkage of palmitate to cysteine residues, which often results in proteins associating with cell membranes60,61. In addition, palmitoylation of the Spike protein is a common strategy for coronaviruses, including SARS-CoV-1 and SARS-CoV-2, to facilitate membrane trafficking and reaching optimal infectivity62. Our data also showed that exogenous palmitate supports viral infection. It is therefore possible that inhibition of FASN could restrict viral infection at multiple levels, including entry as well as viral protein-host membrane interactions. However, future studies are necessary to fully elucidate the mechanisms responsible for the function of FASN in viral entry and other steps in the viral life cycle.
More importantly, our studies indicate that FASN inhibitors can act as broad antiviral agents against different types of viruses, including VSV, HSV-1, MHV68 and especially emerging infectious viruses such as SARS-CoV-2. Many antiviral drugs target specific viral proteins such as the HIV reverse transcriptase or influenza neuraminidase, which have been developed over years of study against such long existing viruses. However, in the past two decades, we have been challenged with numerous emerging and highly pathogenic viruses, including SARS, MERS, Ebola, Zika and SARS-CoV-2. We are still learning about the pathogenesis of these viruses, and we do not always have the luxury of waiting for decades of research to show us the optimal drug targets. Agents that apply broadly-acting antiviral strategies could potentially be used as first line agents against novel diseases caused by many different types of viruses. We previously demonstrated that 25-hydroxycholesterol (25HC), the metabolic product of the IFN-I inducible cholesterol 25-hydroxylase, is a broad-spectrum antiviral agent against numerous types of viruses, including SARS-CoV-263. In this manuscript, we provide evidence that inhibitors of FASN can also be used as broad-spectrum antiviral agents. In particular, we have shown that multiple FASN inhibitors, including C75, EGCG, Cerulenin and TVB-3166, could strongly suppress SARS-CoV-2 infection. Additionally, downregulation or knockdown of Fasn also results in inhibiting SARS-CoV-2 infection. We have also provided evidence suggesting that the FASN inhibitors can suppress infection by different SARS-CoV-2 variants, including α, β, γ and δ SARS-CoV-2 at a similar efficacy as the WT SARS-CoV-2.
Interestingly, AM-580, an inhibitor of sterol regulatory element binding protein-1 (SREBP-1) has been shown to have antiviral properties against MERS-CoV in Huh7.5 cells64. We have previously shown that SARS-CoV-2 stabilizes the SREBP-1 by protease cleavage of IFN-I inducible gene, ring finger protein 20 (RNF-20) for optimum infection65. Thus, future studies should explore the role of SREBP-1 inhibition on viral infection through regulation of its downstream gene, FASN. We have shown that IFN-I downregulates FASN as a defense mechanism against wide range of enveloped viruses. Overall, our findings uncover a novel mechanism for IFN-I in host defense against viral infection by suppressing FASN, which catalyzes fatty acid synthesis. Besides Fasn, we found that other lipid metabolic genes such as Acaca and Acsl3 are also suppressed by IFN-I. As all viruses take advantage of host metabolic products for their rapid growth, suppressing host metabolic genes might be a general and effective strategy to control viral growth. Further studies in the cross-talk between innate immune pathways and host metabolisms may provide new insights in developing broad spectrum antiviral agents against different types of viral infections and resulting diseases like COVID-19.
5. Conclusions
Viruses reprogram the cellular metabolism of the host cell for optimal growth. The host innate immune signaling pathway counteracts viral invasion by inducing type-I-interferon (IFN-I). Subsequently, IFN-I up-/downregulates expression of particular genes to block viral infection. Fatty acid synthase (FASN), the sole enzyme involved in de novo fatty acid synthesis is downregulated possibly by IFN-I upon viral infection. We have shown that FASN deficiency or suppression by pharmacological inhibitors including C75, cerulenin, EGCG, and TVB-3166 significantly blocks replication and spread of a wide variety of enveloped viruses including SARS-CoV-2 and its variants of concern. As viruses rely entirely on host cell's molecular machinery and metabolic products for adequate growth, suppressing host metabolic genes might be a general and effective strategy to fight against novel diseases caused by many different types of viruses.
Acknowledgments
This project is supported by the Research Funds from US National Institute of Health funds (AI069120, AI158154, and AI149718), the UCLA AIDS Institute and UCLA David Geffen School of Medicine—Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Award Program and Tumor Immunology Training Grant (T32CA912036A1, USA). We thank Dr. Vaithilingarajai Arumugaswami at UCLA for sharing the SARS-CoV-2 variants of concerns obtained from the BEI resources. We thank Ms. Barbara Dillon, the director of the UCLA BSL3 facility for providing a safe and organized laboratory for performing the SARS-CoV-2-based research projects.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.02.019.
Author contributions
Saba R. Aliyari and Amir Ali Ghaffari contributed equally in the design of the study and writing of this manuscript. Amir Ali Ghaffari started the project and was joined later with Saba R. Aliyari in the design and execution of the majority of the experiments. Olivier Pernet in Benhur Lee lab assisted in the design and performance of membrane fusion studies and with Armin Takallou and Adele Zhang generated CRISPR-based FASN knockout cells. Kislay Parvatiyar assisted with experimental design. Clay F. Semenkovich, and Xiaochao Wei assisted with generating LysM-Fasn mice. Karen Reue and Laurent Vergnes provided reagents and valuable expertise in metabolism. Linda D. Chilin and Yuntao Wu designed and performed the Ha-CoV-2 pseudo-virus inhibition assays. Ann-Jay Tong from Stephen Smale laboratory contributed to the performance of RNAseq. Yao Wang and Hoda Gerami assisted in performing gene regulation studies and all authors read and approved the manuscript. Genhong Cheng is involved in overall project design and manuscript preparation.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Suppl fig S1.
Suppl fig S2.
Suppl fig S3.
Suppl fig S4.
Suppl fig S5.
Suppl fig S6.
Suppl fig S7.
Suppl fig S8.
Suppl fig S9.
Suppl fig S10.
References
- 1.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. https://www.Who.Int/Emergencies/Diseases/Novel-Coronavirus-2019.
- 4.Henderson L.A., Canna S.W., Friedman K.G., Gorelik M., Lapidus S.K., Bassiri H., et al. American college of rheumatology clinical guidance for multisystem inflammatory syndrome in children associated with SARS-CoV-2 and hyperinflammation in pediatric COVID-19: version 2. Arthritis Rheumatol. 2021;73:e13–29. doi: 10.1002/art.41616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akira S. Pathogen recognition by innate immunity and its signaling. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:143–156. doi: 10.2183/pjab.85.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fensterl V., Sen G.C. Interferons and viral infections. Biofactors. 2009;35:14–20. doi: 10.1002/biof.6. [DOI] [PubMed] [Google Scholar]
- 7.Janeway C.A., Jr., Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 8.Meylan E., Tschopp J., Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 9.Seth R.B., Sun L., Chen Z.J. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- 10.Akira S., Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa H., Barber G.N. Sting is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai T., Akira S. Antiviral signaling through pattern recognition receptors. J Biochem. 2007;141:137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- 14.Toshchakov V., Jones B.W., Perera P.Y., Thomas K., Cody M.J., Zhang S., et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 15.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 16.Chelbi-Alix M.K., Wietzerbin J. Interferon, a growing cytokine family: 50 years of interferon research. Biochimie. 2007;89:713–718. doi: 10.1016/j.biochi.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Le Page C., Génin P., Baines M.G., Hiscott J. Interferon activation and innate immunity. Rev Immunogenet. 2000;2:374–386. [PubMed] [Google Scholar]
- 18.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S.Y., Sanchez D.J., Aliyari R., Lu S., Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S.Y., Aliyari R., Chikere K., Li G., Marsden M.D., Smith J.K., et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Der S.D., Zhou A., Williams B.R., Silverman R.H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Megger D.A., Philipp J., Le-Trilling V.T.K., Sitek B., Trilling M. Deciphering of the human interferon-regulated proteome by mass spectrometry-based quantitative analysis reveals extent and dynamics of protein induction and repression. Front Immunol. 2017;8:1139. doi: 10.3389/fimmu.2017.01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer C.M., Schafer X.L., Moorman N.J., Munger J. Human cytomegalovirus induces the activity and expression of acetyl-coenzyme a carboxylase, a fatty acid biosynthetic enzyme whose inhibition attenuates viral replication. J Virol. 2011;85:5814–5824. doi: 10.1128/JVI.02630-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez E.L., Pulliam T.H., Dimaio T.A., Thalhofer A.B., Delgado T., Lagunoff M. Glycolysis, glutaminolysis, and fatty acid synthesis are required for distinct stages of Kaposi's sarcoma-associated herpesvirus lytic replication. J Virol. 2017;91 doi: 10.1128/JVI.02237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heaton N.S., Perera R., Berger K.L., Khadka S., Lacount D.J., Kuhn R.J., et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tongluan N., Ramphan S., Wintachai P., Jaresitthikunchai J., Khongwichit S., Wikan N., et al. Involvement of fatty acid synthase in Dengue virus infection. Virol J. 2017;14:28. doi: 10.1186/s12985-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang W.C., Lin R.J., Liao C.L., Lin Y.L. Rab 18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication. J Virol. 2014;88:6793–6804. doi: 10.1128/JVI.00045-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang N., Zhao H., Zhang L. Fatty acid synthase promotes the palmitoylation of chikungunya virus NSP1. J Virol. 2019;93 doi: 10.1128/JVI.01747-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaunt E.R., Cheung W., Richards J.E., Lever A., Desselberger U. Inhibition of rotavirus replication by downregulation of fatty acid synthesis. J Gen Virol. 2013;94:1310–1317. doi: 10.1099/vir.0.050146-0. [DOI] [PubMed] [Google Scholar]
- 30.Aragonès G., Alonso-Villaverde C., Oliveras-Ferraros C., Beltrán-Debón R., Rull A., Rodríguez-Sanabria F., et al. Infection with HIV and HCV enhances the release of fatty acid synthase into circulation: evidence for a novel indicator of viral infection. BMC Gastroenterol. 2010;10:92. doi: 10.1186/1471-230X-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J.T., Tseng C.P., Liao M.H., Lu S.C., Yeh W.Z., Sakamoto N., et al. Hepatitis C virus replication is modulated by the interaction of nonstructural protein NS5B and fatty acid synthase. J Virol. 2013;87:4994–5004. doi: 10.1128/JVI.02526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W., Hood B.L., Chadwick S.L., Liu S., Watkins S.C., Luo G., et al. Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production. Hepatology. 2008;48:1396–1403. doi: 10.1002/hep.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei X., Song H., Yin L., Rizzo M.G., Sidhu R., Covey D.F., et al. Fatty acid synthesis configures the plasma membrane for inflammation in diabetes. Nature. 2016;539:294–298. doi: 10.1038/nature20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian X., Xu F., Lung W.Y., Meyerson C., Ghaffari A.A., Cheng G., et al. Poly I:C enhances susceptibility to secondary pulmonary infections by Gram-positive bacteria. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Wang S., Zhu H., Zheng C. Cloning of the herpes simplex virus type 1 genome as a novel luciferase-tagged infectious bacterial artificial chromosome. Arch Virol. 2011;156:2267–2272. doi: 10.1007/s00705-011-1094-9. [DOI] [PubMed] [Google Scholar]
- 36.Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatt A.P., Jacobs S.R., Freemerman A.J., Makowski L., Rathmell J.C., Dittmer D.P., et al. Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc Natl Acad Sci U S A. 2012;109:11818–11823. doi: 10.1073/pnas.1205995109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levin J.Z., Yassour M., Adiconis X., Nusbaum C., Thompson D.A., Friedman N., et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C., Pachter L., Salzberg S.L. Tophat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takada A., Robison C., Goto H., Sanchez A., Murti K.G., Whitt M.A., et al. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci U S A. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hetrick B., He S., Chilin L.D., Dabbagh D., Alem F., Narayanan A., et al. Development of a novel hybrid alphavirus-SARS-CoV-2 particle for rapid in vitro screening and quantification of neutralization antibodies, antiviral drugs, and viral mutations. bioRxive. 2021 doi: 10.1101/2020.12.22.423965. Available from . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf M.C., Wang Y., Freiberg A.N., Aguilar H.C., Holbrook M.R., Lee B. A catalytically and genetically optimized beta-lactamase-matrix based assay for sensitive, specific, and higher throughput analysis of native henipavirus entry characteristics. Virol J. 2009;6:119. doi: 10.1186/1743-422X-6-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhajda F.P., Pizer E.S., Li J.N., Mani N.S., Frehywot G.L., Townsend C.A. Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proc Natl Acad Sci U S A. 2000;97:3450–3454. doi: 10.1073/pnas.050582897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian W.X. Inhibition of fatty acid synthase by polyphenols. Curr Med Chem. 2006;13:967–977. doi: 10.2174/092986706776361012. [DOI] [PubMed] [Google Scholar]
- 45.Deepa P.R., Vandhana S., Muthukumaran S., Umashankar V., Jayanthi U., Krishnakumar S. Chemical inhibition of fatty acid synthase: molecular docking analysis and biochemical validation in ocular cancer cells. J Ocul Biol Dis Inf. 2010;3:117–128. doi: 10.1007/s12177-011-9065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falchook G., Infante J., Arkenau H.T., Patel M.R., Dean E., Borazanci E., et al. First-in-human study of the safety, pharmacokinetics, and pharmacodynamics of first-in-class fatty acid synthase inhibitor TVB-2640 alone and with a taxane in advanced tumors. EClinicalMedicine. 2021;34:100797. doi: 10.1016/j.eclinm.2021.100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munger J., Bennett B.D., Parikh A., Feng X.J., McArdle J., Rabitz H.A., et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vastag L., Koyuncu E., Grady S.L., Shenk T.E., Rabinowitz J.D. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shavinskaya A., Boulant S., Penin F., McLauchlan J., Bartenschlager R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem. 2007;282:37158–37169. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- 50.Delgado T., Sanchez E.L., Camarda R., Lagunoff M. Global metabolic profiling of infection by an oncogenic virus: KSHV induces and requires lipogenesis for survival of latent infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martín-Acebes M.A., Blázquez A.B., Jiménez de Oya N., Escribano-Romero E., Saiz J.C. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Q., Zhou L., Sun X., Yan Z., Hu C., Wu J., et al. Altered lipid metabolism in recovered sars patients twelve years after infection. Sci Rep. 2017;7:9110. doi: 10.1038/s41598-017-09536-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghislain J.J., Wong T., Nguyen M., Fish E.N. The interferon-inducible STAT2:STAT1 heterodimer preferentially binds in vitro to a consensus element found in the promoters of a subset of interferon-stimulated genes. J Interferon Cytokine Res. 2001;21:379–388. doi: 10.1089/107999001750277853. [DOI] [PubMed] [Google Scholar]
- 54.Wesoly J., Szweykowska-Kulinska Z., Bluyssen H.A. STAT activation and differential complex formation dictate selectivity of interferon responses. Acta Biochim Pol. 2007;54:27–38. [PubMed] [Google Scholar]
- 55.Li C., Deng Y.Q., Wang S., Ma F., Aliyari R., Huang X.Y., et al. 25-Hydroxycholesterol protects host against Zika virus infection and its associated microcephaly in a mouse model. Immunity. 2017;46:446–456. doi: 10.1016/j.immuni.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Armas-Rillo L., Valera M.S., Marrero-Hernández S., Valenzuela-Fernández A. Membrane dynamics associated with viral infection. Rev Med Virol. 2016;26:146–160. doi: 10.1002/rmv.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heaton N.S., Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19:368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazzon M., Mercer J. Lipid interactions during virus entry and infection. Cell Microbiol. 2014;16:1493–1502. doi: 10.1111/cmi.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gouttenoire J., Pollán A., Abrami L., Oechslin N., Mauron J., Matter M., et al. Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M., et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sobocińska J., Roszczenko-Jasińska P., Ciesielska A., Kwiatkowska K. Protein palmitoylation and its role in bacterial and viral infections. Front Immunol. 2017;8:2003. doi: 10.3389/fimmu.2017.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Z., Zhang Z., Wang X., Zhang J., Ren C., Li Y., et al. Palmitoylation of SARS-CoV-2 S protein is essential for viral infectivity. Signal Transduct Target Ther. 2021;6:231. doi: 10.1038/s41392-021-00651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zu S., Deng Y.Q., Zhou C., Li J., Li L., Chen Q., et al. 25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor. Cell Res. 2020;30:1043–1045. doi: 10.1038/s41422-020-00398-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan S., Chu H., Chan J.F., Ye Z.W., Wen L., Yan B., et al. Srebp-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat Commun. 2019;10:120. doi: 10.1038/s41467-018-08015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang S., Wang J., Cheng G. Protease cleavage of RNF20 facilitates coronavirus replication via stabilization of SREBP1. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2107108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.