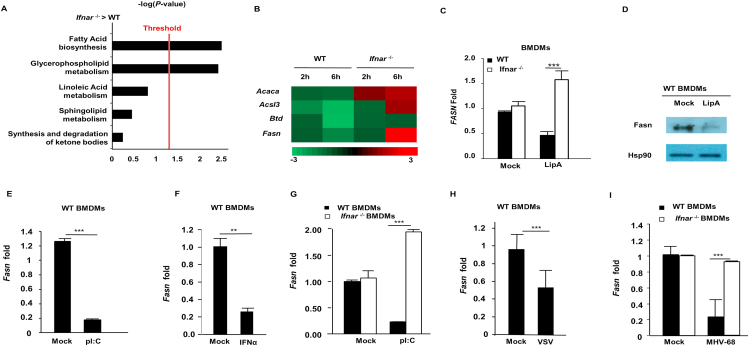
Figure 1.
IFN-I downregulates FASN expression through IFNAR/STAT1 pathways. (A) BMDMs isolated from WT or Ifnar–/– mice were stimulated with LipA (100 ng/mL) for 6 h and RNA samples were isolated and prepared for RNAseq analysis. Using Ingenuity program, canonical pathways which were differentially modulated in Ifnar–/–vs. WT BMDMs were identified. (B) Heat map of fold changes in genes significantly modulated in fatty acid biosynthesis based on the RNAseq data. (C) BMDMs derived from WT and Ifnar–/– mice stimulated with 200 ng/mL LipA and Fasn mRNA levels were assessed by RT-qPCR analysis. (D) WT BMDMs were treated with LipA (200 ng/mL) for 6 h and Fasn protein level was measured by Western blotting. (E) Quantification of Fasn mRNA in WT BMDMs treated with 200 ng/mL pI:C by RT-qPCR. (F) WT BMDMs were treated with IFNα (1000 U/mL) for 6 h and Fasn mRNA was measured by RT-qPCR. (G) Fasn mRNA was quantified in WT and Ifnar–/– BMDMs treated with pI:C (200 ng/mL). (H) The level of Fasn mRNA was measured in WT BMDMs infected with VSV-GFP (MOI = 1) for 8 h. (I) Fasn mRNA was measured in WT and Ifnar–/– BMDMs infected with MHV-68 (MOI = 1) for 8 h. Data presented as mean + SD, n = 3; ∗∗P < 0.01, ∗∗∗P < 0.001.