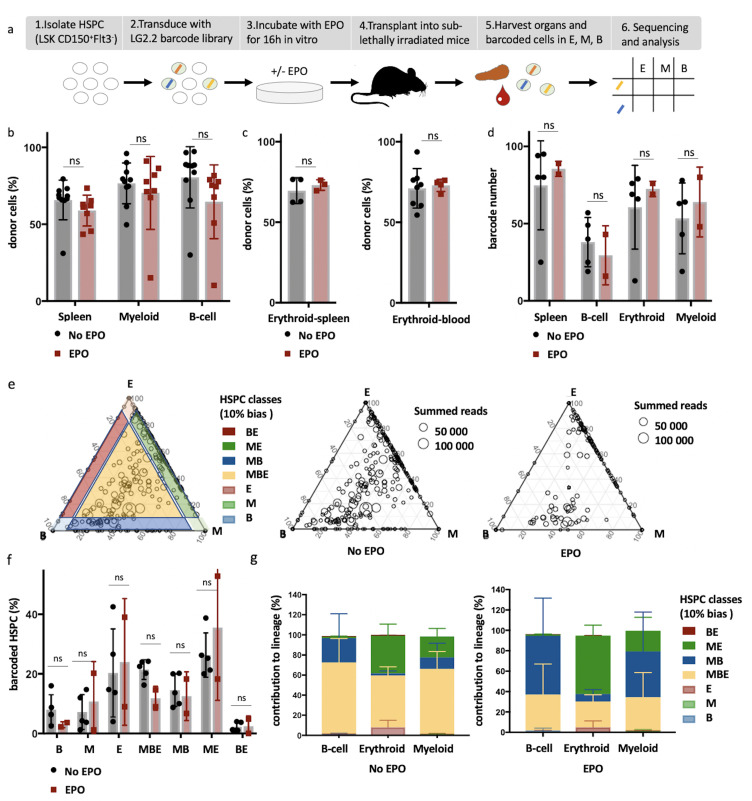
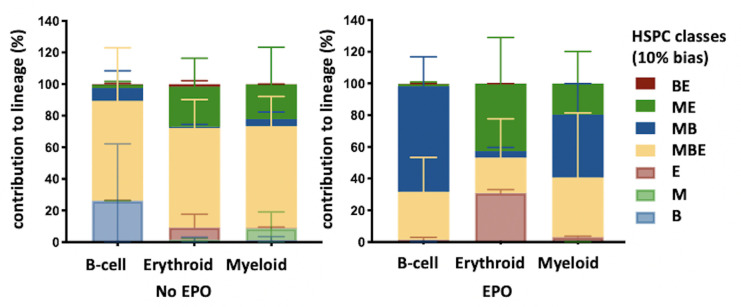
Figure 1. High-output ME- and MB-biased clones occur after erythropoietin (EPO) exposure and transplantation of hematopoietic stem and progenitor cells (HSPCs).
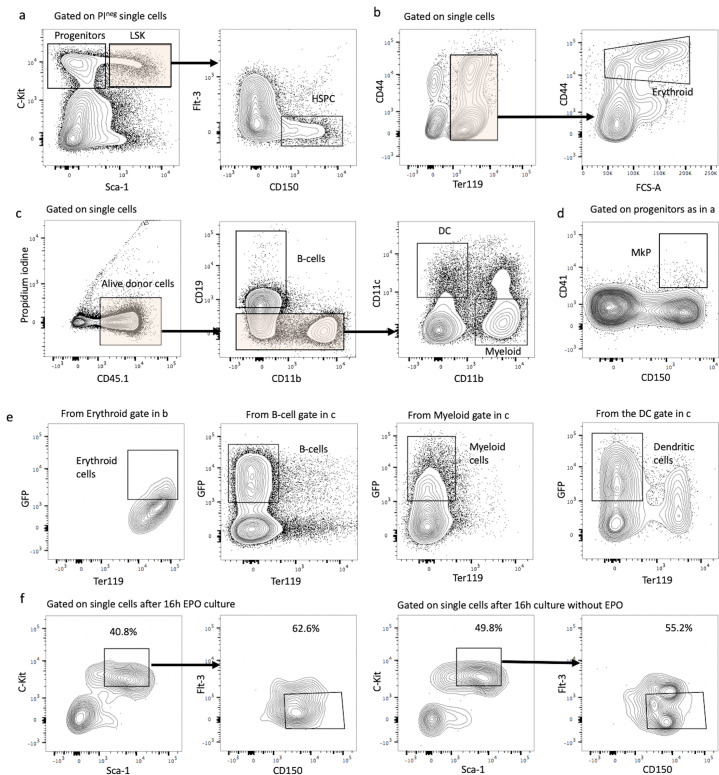
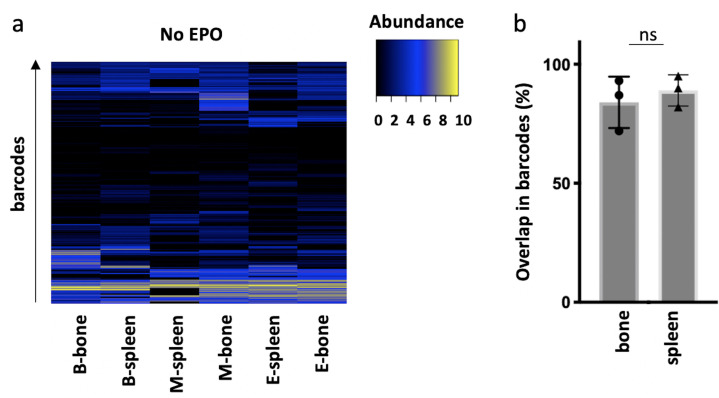
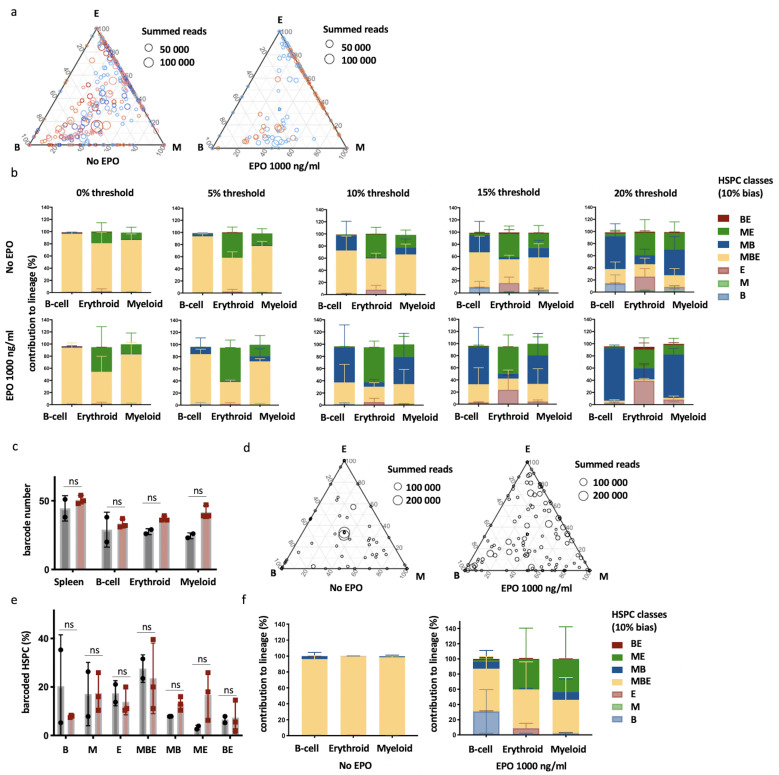
(a) HSPCs were sorted from the bone marrow of donor mice, lentivirally barcoded, cultured ex vivo with or without 1000 ng/ml EPO for 16 hr, and transplanted into sublethally irradiated mice. At week 4 post-transplantation, the erythroid (E), myeloid (M), and B-cells (B) lineages were sorted from the spleen and processed for barcode analysis. (b) The percentage of donor-derived cells (CD45.1+) among the total spleen, myeloid cells (CD11b+) or B-cells (CD19+) in the spleen of control and EPO group. (c) To better assess chimerism in erythroid cells, mTdTomato/mGFP donor mice were used. The fraction of Tom+ cells among erythroid cells (Ter119+) in the spleen and blood in control and EPO group. (d) The number of barcodes retrieved in the indicated lineages at week 4 after transplantation in the control and EPO groups. (e) Triangle plots showing the relative abundance of barcodes (circles) in the E, M, and B lineage with respect to the summed output over the three lineages (size of the circles) for the control and EPO groups. (f) Tthe percentage of HSPCs classified by the indicated lineage bias using a 10% threshold for categorization. (g) Quantitative contribution of the classes as in (f) to each lineage. Shown are values from several animals (n = 8 EPO, n = 10 control in b, n = 3 EPO, n = 4 control in c , spleen, n = 4 EPO, n = 8 control in c , blood collected over five different experiments d–g, n = 5 for the control group and n = 2 for the EPO group collected over one experiment). For all bar graphs, mean and SD between mice are depicted. Statistical significance tested using Mann–Whitney U-test p=0,05 for (b, c). Statistical significance tested by permutation test for different subsets in (g) (see Table 1).