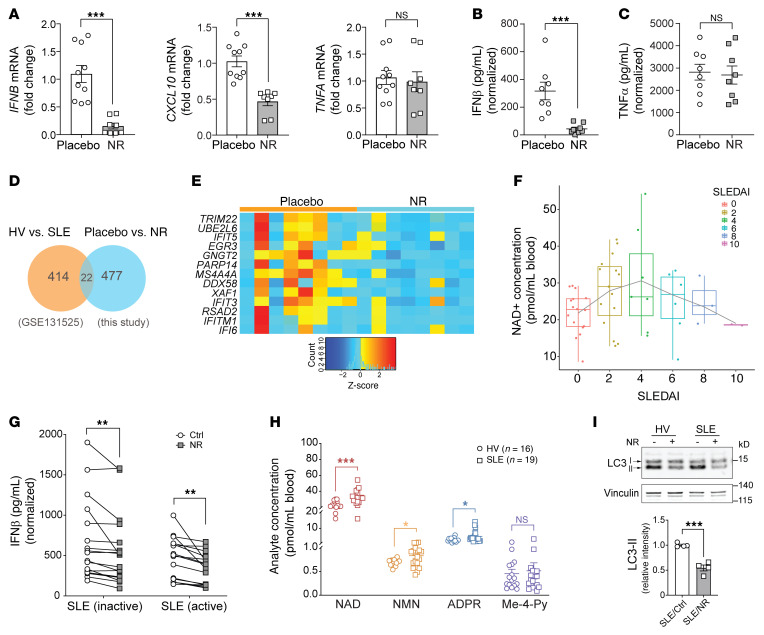
Figure 8. NR blunts monocytic type I IFN in healthy volunteers and in monocytes from patients with SLE.
(A) Relative mRNA expression of IFNB, CXCL10, and TNFA in cryopreserved monocytes from healthy volunteers exposed to the placebo or NR for 7 days (n = 8–10 individuals/group). (B) IFN-β and (C) TNF-α production measured by ELISA in parallel from cryopreserved monocytes described in Figure 7A (n = 8 individuals/group). (D) Venn diagram showing 22 overlapping DE genes between controls versus SLE (GSE131525) and placebo versus NR comparison (this study). (E) Heatmap on relative levels of 13 overlapping DE genes that were downregulated by NR. (F) A box plot of NAD+ concentration for the indicated SLEDAI score on the x axis is depicted. The dot/circle on each box plot represents individual NAD+ measurement for the indicated SLEDAI score. A trend line was generated using LOESS (locally estimated scatter plot smoothing). (G) IFN-β production induced by LPS (10 ng/mL for 6 hours) was measured by ELISA in monocytes from patients with SLE with different disease activity (inactive: SLEDAI ≤4; active: SLEDAI >4) that incubated with vehicle or NR for 16 hours (n = 17 for SLE inactive group; n = 16 for SLE active group). (H) Levels of NAD+, NMN, and ADPR from whole blood of healthy volunteers (HV) and patients with SLE measured by LC-MS (n = 16 for HV group; n = 19 for SLE group). (I) Representative immunoblot and quantification of LC3-II in SLE monocytes incubated with vehicle or NR for 16 hours and then stimulated with LPS (10 ng/mL) for 2 hours. Data were analyzed by unpaired 2-tailed Student’s t test (A–C, H, and I) or paired 2-tailed Student’s t test (G). All data represented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.