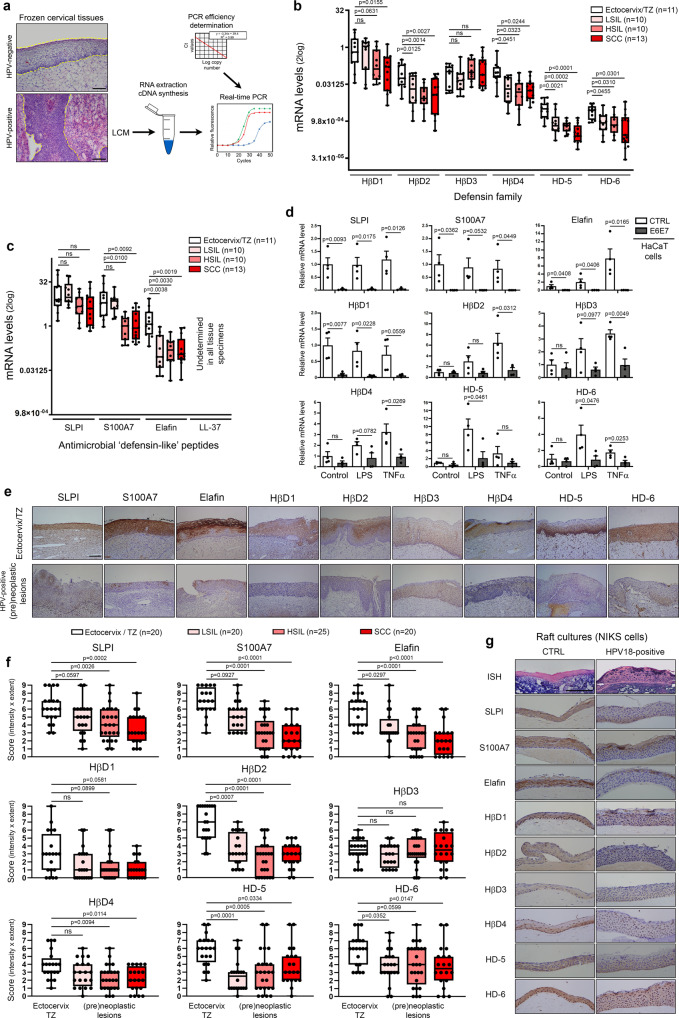
Fig. 2. HPV oncoproteins impair both the constitutive and inducible innate (antimicrobial) peptide expression.
a Schematic illustration of the steps involved in gene expression analysis using microdissected frozen specimens. mRNA expression of epithelial-specific members of the defensin b and “defensin-like” c families was measured by RT-qPCR in HPV-positive (pre)neoplastic lesions (LSIL, n = 10; HSIL, n = 10; SCC, n = 13). Uninfected squamous samples [ectocervix, vagina, transformation zone (TZ)] (n = 11) were used as control. Gene expression was normalized using four calibrator genes (HPRT, GAPDH, 18S and TBP). Box limits: 25th to 75th percentiles; line: median; whiskers: minimum to maximum. d RT-qPCR analysis of SLPI, S100A7, elafin, HβD1-4 and HD-5/6 expression in immortalized keratinocytes stably transduced or not with HPV16 E6/E7 oncoproteins and stimulated with TNFα or LPS. Each experiment was normalized to the amount of HPRT mRNA from the same sample. Results represent the means ± SEM of four independent experiments. e HPV-negative and positive tissue specimens stained for antimicrobial peptides expressed by the squamous epithelium lining the lower part of the female reproductive tract. A reduced immunoreactivity for most of these innate factors was clearly observed in HPV-infected (pre)neoplastic lesions. f Semi-quantitative evaluation of innate peptide expression (intensity and extent of the immunostainings) in both normal epithelium from HPV-negative samples (n = 20) and (pre)neoplastic lesions (LSIL, n = 20; HSIL, n = 25; SCC, n = 20). Box limits: 25th to 75th percentiles; line: median; whiskers: minimum to maximum. g Representative control and HPV18-positive organotypic raft culture sections stained for all analyzed host defense (antimicrobial) peptides. As a control, HPV DNA was detected by in situ hybridization and, consistent with an episomal infection, a diffuse punctate pattern was observed. Images are representative of three independent experiments. The scale bar represents 100 μm. P values were determined using one-way ANOVA followed by Dunnett’s multiple comparison post-hoc test b, c, two-sided unpaired t-tests d and ANOVA Kruskal–Wallis test followed by Dunn post-hoc test f. ns: not significant (p > 0.1). Source data are provided as a Source Data file.