Figure EV5. Additional analysis of the translation elongation rate in zebrafish embryos.
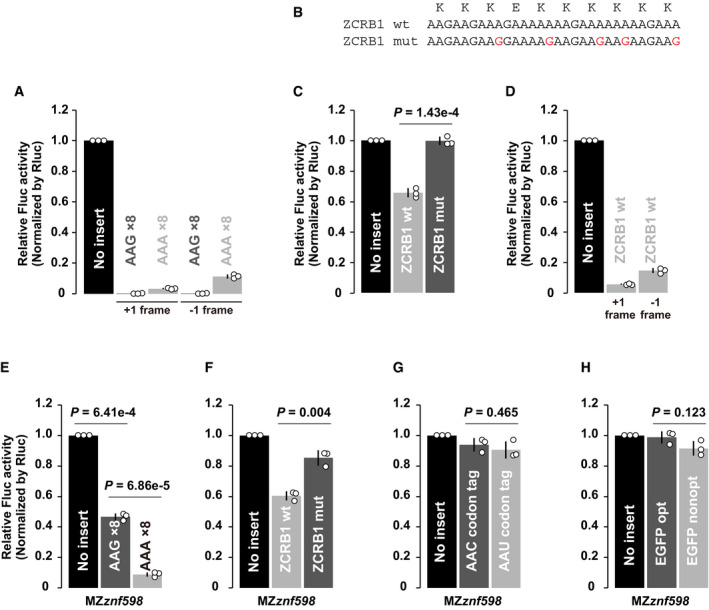
- Results of the tandem ORF assay to detect frameshifting with Lys AAG×8 or Lys AAA×8 sequences. One or two guanine nucleotides (G or GG) were inserted after Lys AAG×8 or Lys AAA×8 sequences to detect protein products from +1 or −1 frame.
- Wild‐type (wt) or mutated (mut) stalling sequences of human ZCRB1. Mutated nucleotides were indicated in red. The amino acid sequence is shown above.
- Results of the tandem ORF assay with wild‐type (wt) or mutated (mut) stalling sequences of ZCRB1.
- Results of the tandem ORF assay to detect frameshifting with a ZCRB1‐stalling sequence. One or two guanine nucleotides (G or GG) were inserted after the ZCRB1 sequence to detect protein products from +1 or −1 frame.
- Results of the tandem ORF assay with Lys AAG×8 or Lys AAA×8 sequences in MZznf598 embryos.
- Results of the tandem ORF assay with wild‐type (wt) or mutated (mut) stalling sequences of ZCRB1 in MZznf598 embryos.
- Results of the tandem ORF assay with Asn AAC and Asn AAU codon tags in MZznf598 embryos.
- Results of the tandem ORF assay using EGFP ORFs with different codon optimalities in MZznf598 embryos.
Data information: In all experiments, normalized Fluc activity with no insert was set to one. All experiments were repeated three times, and the average Fluc activity is shown as bar charts. Error bars show SD. Individual data points are shown as dots. P‐values were calculated using two‐sided Student’s t‐test.
Source data are available online for this figure.
