Molecular mechanistic biology has ushered us into the world of life’s building blocks, revealing their interactions in macromolecular complexes and inspiring strategies for detailed functional interrogations. The biogenesis of membraneless cellular compartments, functional mesoscale subcellular locales devoid of strong internal order and delimiting membranes, is among mechanistic biology’s most demanding current challenges. A developing paradigm, biomolecular phase separation, emphasizes solvation of the building blocks through low‐affinity, weakly adhesive unspecific interactions as the driver of biogenesis of membraneless compartments. Here, I discuss the molecular underpinnings of the phase separation paradigm and demonstrate that validating its assumptions is much more challenging than hitherto appreciated. I also discuss that highly specific interactions, rather than unspecific ones, appear to be the main driver of biogenesis of subcellular compartments, while phase separation may be harnessed locally in selected instances to generate material properties tailored for specific functions, as exemplified by nucleocytoplasmic transport.
Subject Categories: Organelles, Structural Biology
This commentary contrasts the contributions of site‐specific versus unspecific biomolecular interactions to the molecular mechanisms of subcellular compartment assembly and function.
Glossary
- CPC
chromosomal passenger complex
- C sat
saturating concentration
- FRAP
fluorescence recovery after photobleaching
- IDP
intrinsically disordered protein
- IDR
intrinsically disordered regions
- KD
dissociation constant
- k off
dissociation rate constant
- MVP
multivalent protein
- NoLS
nucleolar localization signal
- NOR
nucleolar organizer region
- NPC
nuclear pore complex
- NTR
nuclear transport receptor
- Nups
nucleoporins
- PRM
proline‐rich motif
- PS
phase separation
- RNP
ribonucleoprotein
- SAC
spindle assembly checkpoint
- SSI
site‐specific interaction
Introduction
Many functions of eukaryotic cells are confined within compartments that are not delimited by membranes (and therefore often referred to as membraneless). Examples that have been known for many decades, among a number of others, are the centrosome, the nucleolus, and P‐granules (Courchaine et al, 2016; Banani et al, 2017; Shin & Brangwynne, 2017; Ditlev et al, 2018; Lyon et al, 2021). With linear dimensions in the biological mesoscale (between ~100 nm and a few micrometers), compartments are supra‐molecular, i.e., larger than their individual macromolecular components. Typically, tens or even hundreds of different macromolecular species, usually in multiple copies, populate each individual compartment and interact in it, conferring different degrees of internal order on compartments (Wu & Fuxreiter, 2016; Goetz & Mahamid, 2020; Peran & Mittag, 2020; Fare et al, 2021; Korkmazhan et al, 2021).
The physicochemical drivers of the biogenesis, maintenance, and disassembly of compartments, and their ability to concentrate macromolecules without an encapsulating membrane, have attracted considerable interest (Banani et al, 2017; Shin & Brangwynne, 2017). Molecular mechanistic biology, with its focus on the identification, characterization, and visualization of discrete and highly specific interactions of macromolecules, has uncovered how the building blocks of compartments assemble and interact reciprocally (see Fig 1A–E for a few examples) (Jones & Thornton, 1996; Sudha et al, 2014). How the building blocks interact to give rise to mesoscale structures devoid of strong internal order, however, is less well understood and harder to model.
Figure 1. Examples of site‐specific interactions and their combination in MVPs.
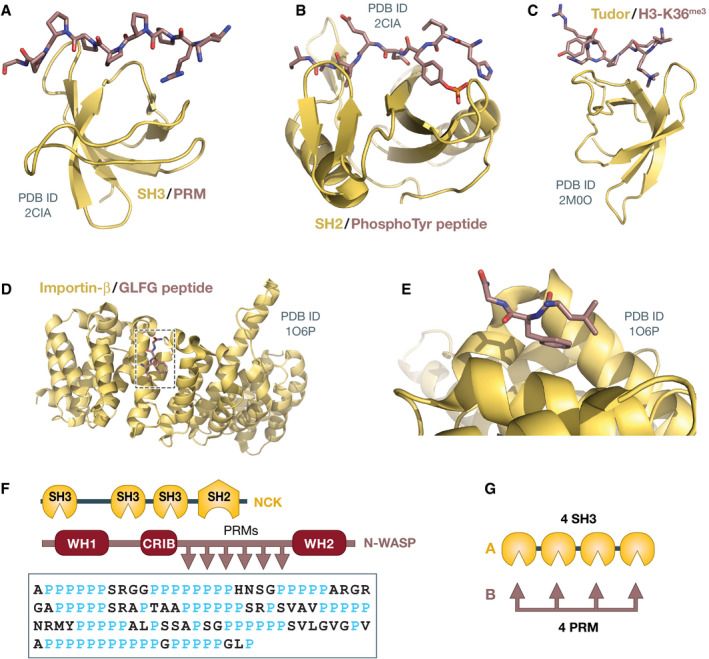
(A–E) Cartoon diagrams of various protein domains and proteins discussed in the text (yellow), with their cognate ligands shown in stick. (A) Src homology 3 (SH3) domain with proline‐rich motif (PRM); (B) Src homology 2 domain with tyrosine‐phosphorylated peptide; (C) Tudor domain with peptide from histone H3 trimethylated on lysine 36; (D) Importin‐β with Gly‐Leu‐Gly‐Phe (GLGF) peptide; (E) Enlargement of area boxed in D. The protein data bank (PDB) codes are indicated; (F) Domain organization of the NCK and N‐WASP proteins, and sequence of the proline‐rich region of N‐WASP (Uniprot, human sequence); and (G) Artificial multivalent constructs used by Li et al (2012).
In recent years, a burgeoning new field of research in cell biology, biomolecular phase separation (PS), has promoted a radically different explanation for how membraneless compartments assemble. Under the PS paradigm, classical specific macromolecular interactions are attributed a secondary role in compartment assembly. Compartments are rather proposed to form under the action of one or a few specific PS drivers (almost invariably proteins). Drawing concepts from polymer chemistry (Hyman et al, 2014; Brangwynne et al, 2015), PS drivers are treated as associative polymers that phase‐separate thanks to transient, low‐affinity, cohesive interactions (Fig 2B and C) (Tanaka, 2011; Hyman et al, 2014; Brangwynne et al, 2015; Uversky et al, 2015; Banani et al, 2017; Shin & Brangwynne, 2017; Boeynaems et al, 2018; Choi et al, 2020).
Figure 2. Examples of IDPs and low‐affinity, non‐site‐specific interactions.
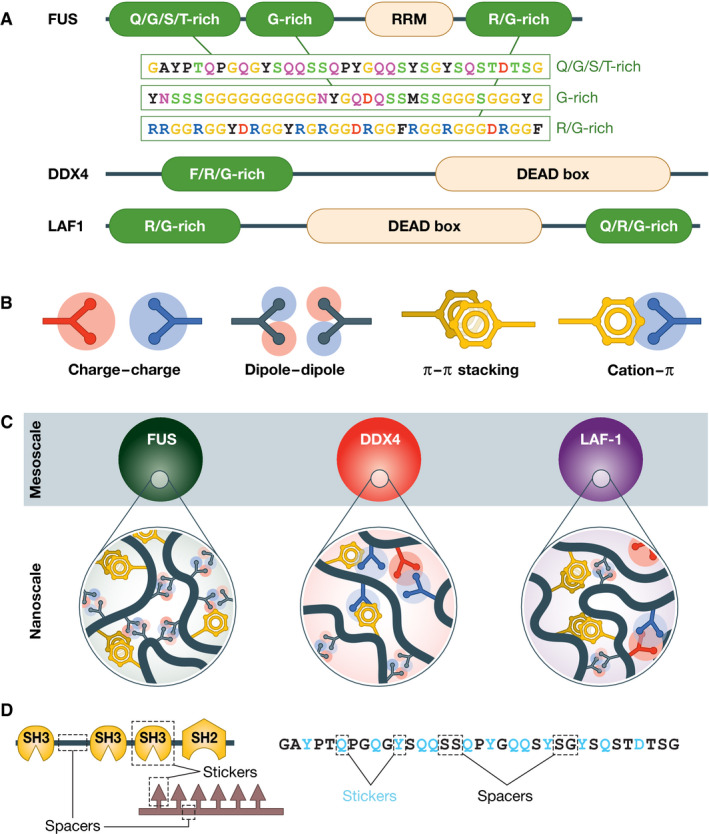
(A) Domain organization of three IDPs and specific sequence stretches from each of three low‐complexity regions of human FUS; (B) Examples of low‐affinity interactions believed to drive homotypic PS of IDPs; (C) Droplets of FUS, DDX4, and LAF‐1 (indicated as “mesoscale”) are proposed to arise from multiple nanoscale interactions shown in B; (D) Sticker‐and‐spacer model for interactions of MVPs (left) and of IDPs (right). Gln and Tyr (Q and Y, respectively) are considered stickers that could interact through dipoles and stacking. The entire figure is an adaptation of Fig 2 from Brangwynne et al (2015).
The PS paradigm proposes that membraneless compartments are (usually) liquid like and that they arise from liquid–liquid (LL) demixing, a process exemplified by the spontaneous, thermodynamically driven separation of oil from water caused by poor solvation (Hyman et al, 2014; Banani et al, 2017). This idea was ignited by provocative observations that P‐granules and nucleoli form droplets that fuse, wet neighboring cellular structures, drip, and display rapid internal mobility of its components when assayed by fluorescence recovery after photobleaching (FRAP) (Phair & Misteli, 2000; Brangwynne et al, 2009, 2011). Subsequent studies on two classes of molecules frequently identified in membraneless compartments, multivalent proteins (MVPs; Fig 1F and G) and intrinsically disordered proteins (IDPs), containing low‐complexity domains (LCDs, examples of which are shown in Fig 2A), began to provide a molecular and theoretical underpinning to the PS paradigm. For instance, natural or engineered MVPs were shown to form droplets in vitro when highly concentrated (Li et al, 2012; Banani et al, 2016; Harmon et al, 2017), and similar observations were also made with various IDPs [e.g., (Kato et al, 2012; Elbaum‐Garfinkle et al, 2015; Lin et al, 2015; Molliex et al, 2015; Nott et al, 2015; Pak et al, 2016)]. These observations supported the progressive identification of MVPs and IDPs as scaffolds driving PS through weak homotypic interactions (i.e., involving the same macromolecule), while other macromolecules attracted to the same compartments through heterotypic interactions (i.e., involving different types of macromolecules) were identified as clients (Hyman & Simons, 2012; Banani et al, 2016, 2017). Collectively, these developments caused the transformation of PS from the abstractions of a useful analogy to a seemingly coherent and unifying paradigm for the biogenesis of membraneless compartments from the nano‐ to the mesoscale (Hyman & Brangwynne, 2011; Banani et al, 2017).
Macromolecular interactions
Below, we will often refer to three main types of macromolecular interactions. The first (type I) are homotypic or heterotypic interactions established (mainly) by IDPs, i.e., by proteins, or segments thereof, that do not adopt a stable folded conformation. IDPs are usually viewed as arrays of stickers and spacers, with spacers determining overall solubility, and stickers mediating interactions (Tompa & Fuxreiter, 2008; Brangwynne et al, 2015; Martin & Mittag, 2018; Choi et al, 2020; Borcherds et al, 2021; Fare et al, 2021). Stickers are capable of only few, relatively low‐affinity and poorly specific types of attractive interactions, including charge–charge, dipole–dipole, cation Π, and Π–Π stacking (Fig 2B).
The second and third types of macromolecular interactions (types II and III) are also homotypic or heterotypic, but are enabled by the specific three‐dimensional arrangement (reflecting conformation) and the detailed chemical identity of the binding interfaces. These interactions involve at least one folded domain of a macromolecule and either (type II) a short linear segment of a target macromolecule, which may also fold locally in the process of binding, or (type III) another folded domain. While type I interactions are mediated by a limited set of attractive or repulsive bonds, with the result that their specificity is limited, macromolecular interactions of types II and III usually involve large, conserved surface patches on the interacting macromolecules that exploit the spatial and chemical complementarity of the binding interfaces as well as steric exclusion (Jones & Thornton, 1996; Aloy et al, 2003; Choi et al, 2009; van der Lee et al, 2014).
The PS idea identifies interactions of type I typically as fuzzy, whereas interactions of types II and III are now often defined stereospecific (Hyman & Brangwynne, 2011; Banani et al, 2017). The term stereospecific is used in chemistry to denote formation, recognition, or reaction of enantiomers. If used with reference to biological interactions, the term stereospecific aims to put emphasis on the property that specificity is largely dictated by the complementary geometry and chemistry of binding sites. This use of stereospecific is therefore related to, but deviates considerably from, the traditional definition of the term. For this reason, here I prefer using simply site‐specific interaction (abbreviated as SSI) to refer to interactions of types II and III.
SSIs are enormously versatile, as masterly described in a classic review (Mammen et al, 1998). As a rule of thumb, they are titrated between 0.1 and 10 times the dissociation constant (KD) (Jarmoskaite et al, 2020). If the KD is sufficiently small (high affinity), correspondingly low concentrations of binders, normally well below their solubility limit, will suffice. SSIs can be easily and rapidly modulated, e.g., through post‐translational modifications, without having to change the concentration of binders. KDs can be extremely low, to the point of making an interaction essentially irreversible, as in the core of many multisubunit macromolecular complexes, or relatively high for regulated interactions, and easily adjustable to non‐equilibrium conditions in active processes. Thus, while in principle, both type I and type II/III interactions can reach high affinity (e.g., for type I interactions by harnessing multivalency), the hallmark that distinguishes I from II and III is the low specificity of the first and the high specificity of the latter two.
Condensates
As discussed below, the narrative that compartments are liquid, and driven by low‐affinity, low‐specificity fuzzy interactions has severe shortcomings. Nonetheless, it has exercised a tremendous influence on the field’s developments. Briefly, after disregarding SSIs as potential drivers of compartment biogenesis on the basis of a supposed incompatibility with the liquid‐like appearance of phase‐separated compartments (Hyman & Brangwynne, 2011), it licensed PS assays in which putative PS drivers were studied in perfect isolation in vitro, and usually confirmed in this presumed role (prematurely, as explained below). This allowed PS, initially invoked for the assembly of nuclear and cytoplasmic bodies involved in ribonucleoprotein (RNP) assembly and metabolism, such as nucleoli and P‐granules, to claim for itself an ever‐growing list of cellular territories. PS is now considered a self‐evident universal driver of compartment assembly, as implied by the proposal to rename all membraneless compartments biomolecular condensates (Banani et al, 2017; Shin & Brangwynne, 2017; Snead & Gladfelter, 2019) on the basis of their ability to concentrate molecules, that they comprise biological macromolecules, and that they may arise through similar mechanisms [text in italic indicates textual citations (Banani et al, 2017)]. This definition encompasses functionally and compositionally diverse compartments like centrosomes, centromeres, kinetochores, transcriptional super‐enhancers, chromatin domains, chromosomes, sites of response to DNA damage and DNA recombination, membrane‐associated signaling complexes (which of course are not “membraneless” per se, but nevertheless not surrounded by a membrane), and many more (Hyman & Brangwynne, 2011; Banani et al, 2017; Shin & Brangwynne, 2017; Lyon et al, 2021).
Not unlike the term stereospecific, also the term condensate, as often used in the PS context, carries some ambiguities. Condensation is usually taken to describe the transition of a gas to a liquid, or chemical bond formation between two molecules accompanied by release of water. In PS publications, the term condensation is rather used to imply concentration of biomolecules through a phase change (Banani et al, 2017)—which may be considered a similar analogy as using condensation to describe the compaction of chromosomes upon mitotic entry. It is important to keep in mind, however, that the ability to concentrate molecules, e.g., to accelerate reactions or inhibit them through sequestration (Banani et al, 2017; Shin & Brangwynne, 2017; Lyon et al, 2021), is an attribute that does not support a role of PS more than it supports other mechanisms of biogenesis. SSIs can do that too, and better, as I will discuss. Furthermore, while concentration of specific macromolecules is clearly a feature of membraneless compartment, whether cellular compartments generally reach overall macromolecular concentrations higher than those of the surrounding cellular medium is unclear (Handwerger et al, 2005; Wei et al, 2017). Thus, collectively, condensate should not be taken as a literal descriptor of mesoscale membraneless compartments, at least in relation to our current knowledge of the processes that promote their assembly.
What drives compartment assembly?
Another question raised by the definition of biomolecular condensates is whether it is true that the similar mechanisms from which compartments are proposed to arise imply homotypic fuzzy interactions of PS scaffolds. These interactions are assumed to be the basis of PS and compartment assembly, but as we shall see, this is rather implausible and defies compelling evidence that the only truly general mechanism of compartment biogenesis involves highly specific heterotypic SSIs occurring at concentrations well below the saturating concentration (C sat) of any of the interacting components, i.e., in the one‐phase regime. These concerns, to the extent that they call into question the very arguments on which the PS concept has built its success, are highly significant and require thorough motivation. In this essay, I will therefore examine two crucial questions: (i) Are low‐specificity, fuzzy interactions of macromolecules as associative polymers literally the primary physicochemical driver of biogenesis of membraneless compartments in cells under physiological conditions (I will refer to this as general PS)? and (ii) Can phase transitions influence a compartment’s solvation and material properties after macromolecules have become concentrated there by more traditional binding mechanisms (I will refer to this as special or restricted PS)? Below I will first discuss arguments indicating that the answer to question 1 is (most) likely no. Later, I will also discuss why the answer to question 2 is instead likely yes, but probably for a relatively small subset of compartments. I will also clarify why demonstrating special PS, let alone general PS (if it exists in our cells), requires standards of proof that studies on PS have almost invariably ignored.
What is general PS?
General PS assumes that low‐specificity homotypic interactions of one or very few PS scaffold macromolecules drive, at concentrations above their C sat, a density transition that results in the formation of coexisting dilute and dense phases, each with a fixed concentration of the scaffold (Banani et al, 2017). The dense phase may further promote concentration of other species to complete assembly of a mature compartment (Hyman et al, 2014; Banani et al, 2017). Examples adapted from Brangwynne et al (2015) are shown in Fig 2C.
As noted, MVPs and IDPs were initially identified as likely scaffolds in general PS. Their plausibility as PS drivers reflected (i) their enrichment in various membraneless compartments and (ii) their tendency, especially for the IDP class, to be aggregation prone and implicated in various degenerative pathologies (Alberti & Hyman, 2021). This ignited an extensive program of study of their phase behavior in vitro, with attempts to correlate it with their behavior in living cells and in disease. For instance, studies focused on the associative properties of tight complexes of reciprocally interacting linear MVPs, including engineered ones containing multiple SH3 domains and proline‐rich motifs (PRMs), separated by flexible linker spacers (Fig 1F and G) (Li et al, 2012; Banani et al, 2016; Harmon et al, 2017; Case et al, 2019).
While IDPs may be viewed as assemblers, whose distributed linear motifs engage in SSIs with target folded domains (type II interactions) (van der Lee et al, 2014), the PS paradigm considers IDPs usually as associative polymers with alternating stickers and spacers (type I) (Choi et al, 2020). The same sticker‐spacer description may be applied to MVPs, where folded domains are stickers‐mediating interactions and the linkers between them are spacers (Choi et al, 2020). This description aims to underline a unifying similarity of MVPs and IDPs that might explain why both classes drive PS (Fig 2D). However, the similarity is less obvious than implied by this unifying description, as discussed more thoroughly below, because the stickers in MVPs usually are SSIs that can generate very significant binding affinities and specificity, whereas the stickers of IDPs are relatively low affinity and non‐specific, at least in non‐aggregative states (Wu & Fuxreiter, 2016; Korkmazhan et al, 2021).
The unintuitive general PS
Fascinating as they may seem, the PS tenets prove on closer scrutiny less intuitive than the simple oil–vinegar analogy suggests. Proteomes may have evolved to maximize specificity and minimize aggregation of proteins (Zhang et al, 2008; Johnson & Hummer, 2011). Neither specificity nor aggregation seem to be of significant concern for proposed general PS mechanisms of condensate biogenesis, which rather focus on low‐affinity and low‐specificity interactions (Fig 2) at or above the solubility limit of one or more PS drivers (Hyman et al, 2014; Brangwynne et al, 2015). It is hard to imagine how fuzzy interactions of very low specificity would promote selective, thermodynamically driven condensation of any particular macromolecule at typical cellular concentrations, with myriad competing interactions of a similar kind. And how can the same limited set of weak, non‐site‐specific interactions explain how different compartments maintain their identities and prohibit or enable co‐PS? Are active processes involved in this complicated scheme, and if so, how? How is the saturating concentration of every putative PS driver adjusted across different specialized cells with different sizes and compositions of their cytosol, and even more so across different organisms? And how would the formation of toxic aggregates be controlled given the proposed generality of this mechanism for compartment biogenesis? Let us also consider that compartments usually form around defined spatial and temporal cues, delimited by the presence of a primary scaffold (Fig 3), and ideally do not extend beyond them. These cues, be it signaling‐active transmembrane receptors or centromeres, attract and concentrate the correct targets almost certainly through SSIs with adequate affinity and specificity, probably also dictating the final concentration of various compartment’s components. If PS drivers were indeed sufficient for PS in the absence of spatial cues, why do they not assemble unrestrained in space and time?
Figure 3. Hierarchy of compartment assembly.
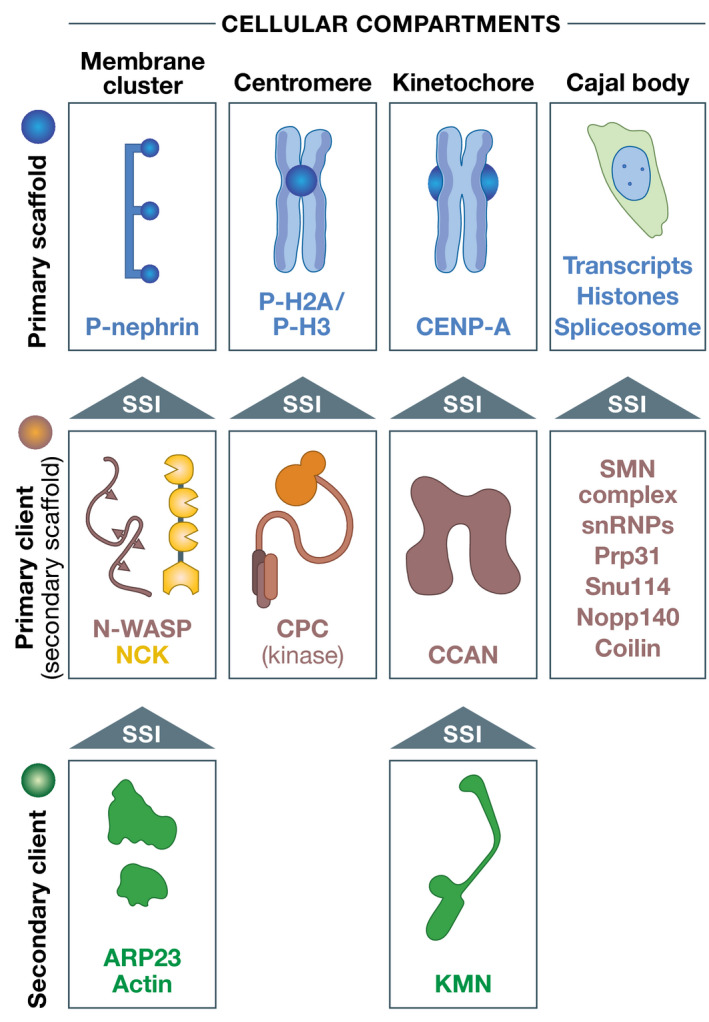
Compartments are usually hierarchical. Nephrin, as an element of membrane clusters, is a transmembrane protein whose intracellular domain undergoes regulated phosphorylation. As primary scaffold, it acts as a binding site for the NCK client through its SH2 domain. N‐WASP binds NCK through phosphorylation. Regulators of actin polymerization may be further recruited to clusters. During mitosis, the CPC kinase complex is recruited to specific phosphorylated residues of histone H2A and H3 that are enriched in the centromere region. It is therefore a client of the centromere, although it has been suggested to act as scaffold in PS. The kinetochore assembles on the histone H3 variant CENP‐A, which is part of a specialized centromeric histone complex. Constitutive centromere‐associated network (CCAN) subunits are recruited through interactions with CENP‐A. The Knl1‐Mis12‐Ndc80 (KMN) complex is recruited to CCAN through established SSIs. Not shown are tertiary clients like the spindle checkpoint proteins BUB1 and MAD2 discussed in the text. In Cajal bodies, specific RNAs transcripts, for instance, of histones, but also spliceosome subunits, act as scaffolds for the recruitment of a variety of downstream proteins (Kaiser et al, 2008; Shevtsov & Dundr, 2011). The recruitment hierarchy remains poorly understood. Other nuclear bodies, like the histone body or the nucleolus, also require transcription (see main text for details).
Liquid and site specific
Thus, general PS may seem to counter biological intuition when it proposes an inverted hierarchy of compartment assembly that prioritizes unspecific interactions over more probable site‐specific ones (Hyman & Brangwynne, 2011). If in all evidence macromolecules usually become concentrated in their target cellular locales due to highly site‐specific interactions, why were these not considered as a mechanism of biogenesis? The primary answer is that they were perceived as incompatible with the liquid‐like behavior of supposed phase‐separated organelles (Hyman & Brangwynne, 2011). For instance, the kinetics of recovery of many components of compartments in FRAP experiments was deemed too fast for SSIs (Hyman & Brangwynne, 2011). This conclusion appears premature. The kinetics of typical SSIs, as they may be captured by half‐time of the bound state at equilibrium, t1/2, are well within boundaries demonstrated by FRAP experiments on liquid‐like compartments [for a more thorough and informative discussion on the execution and interpretation of FRAP experiments, including more sophisticated protocols to assess the potential presence of PS, please consult Erdel et al (2020; McSwiggen et al (2019b); Sprague and McNally (2005); Taylor et al (2019) and references therein]. If a macromolecule bound to a receptor in a compartment (rather than getting there through condensation), its FRAP recovery rates would be typically determined by the dissociation rate constant (k off), i.e., by the rate of release of the bleached molecules from their receptor, necessary for replacement with new fluorescent molecules (Sprague & McNally, 2005). The faster the bleached molecules dissociate from their receptor, the faster the recovery.
For example, the spindle assembly checkpoint (SAC) complex BUB1/BUB3 binds its kinetochore receptor with a dissociation constant (KD) of approximately 100 nM and a t1/2 of recovery in FRAP experiments of ~10 s (Overlack et al, 2015, 2017) (Fig 4A and B). Similarly, FRAP recovery rates and fractions for another SAC protein at kinetochores, MAD2, were initially measured in living cells and then reproduced with recombinant proteins in a reconstituted system in vitro, with typical t1/2 of recovery of a few seconds (Howell et al, 2004; Shah et al, 2004; Vink et al, 2006). Other kinetochore components (indicated as “core kinetochore” in Fig 4A) remain connected to the underlying chromatin throughout a cell’s life, without ever exchanging (Jansen et al, 2007). At kinetochores, both rapid and slow/non‐existent turnovers reflect SSIs (Fig 4) (Musacchio & Desai, 2017).
Figure 4. Liquid like does not exclude SSIs.
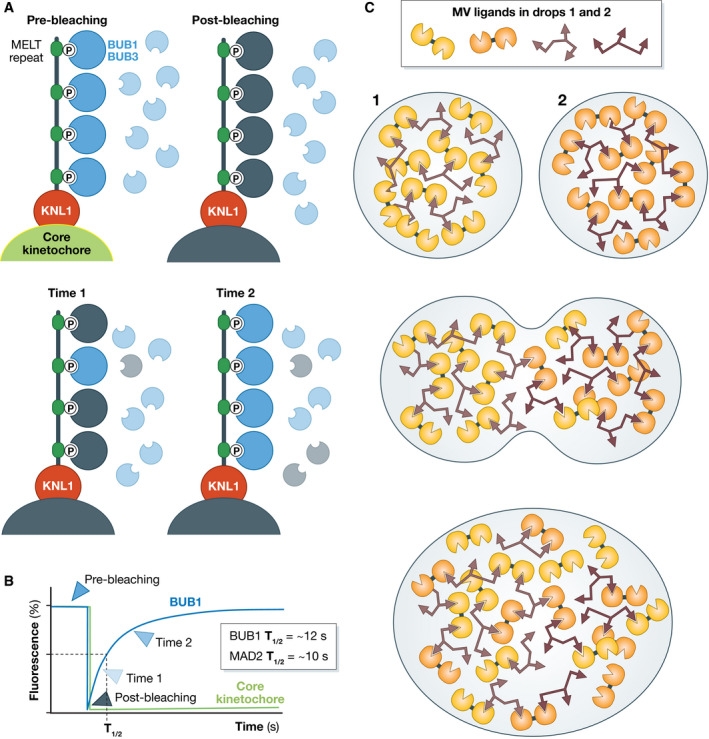
(A) KNL1, an IDP at kinetochores, contains multiple sequence‐related phosphorylation sites for the recruitment of the BUB1/BUB3 complex, where BUB3 is a phospho–amino acid adaptor. KNL1 docks to CCAN in the core kinetochore. A FRAP experiment on BUB1/BUB3 and on a core kinetochore subunit would lead to fundamentally different conclusions on the nature of the compartment, as the core subunit do not exchange and would not recover, whereas BUB1/BUB3 would exchange in seconds; (B) Hand‐drawn curves representing the recovery behavior shown in A; (C) Two imaginary directly interacting MVPs in neighboring phase‐separated droplets (1 and 2, where the small differences in color are meant to recognize the origin of molecules in the original droplets) could easily mix if the interaction times of the individual modules allowed relatively rapid exchange.
In FRAP experiments, predicted recovery half‐times of macromolecular interactions, just assuming typical KDs for reversible interactions and typical association rate constants (Shammas et al, 2012), will thus extend from sub‐seconds to hours, considered to indicate a diffusive liquid‐like state or even solid‐like state in many claims of PS [typically 1–100 s (McSwiggen et al, 2019b)]. If SSIs turn over rapidly, as they can, they provide a plausible basis for another property of liquid‐like membraneless compartment, their fusion. Turning over rapidly, new “mixed” interactions in fusing droplets will progressively replace the identical interactions that existed in the individual droplets, especially if the network is heavily cross‐linked through interactions turning over rapidly, as shown in a hypothetical example in Fig 4C [see also discussion in (Erdel & Rippe, 2018)]. In cells, active processes may additionally contribute to promote fusions, as observed for mitotic spindles, large ensembles of microtubules, microtubule cross‐linkers, and molecular motors engaged in many reciprocal dynamic SSIs, which can fuse within a few minutes (Gatlin et al, 2009).
Finally, how surface tension/energy of membraneless compartments emerges, and which compartments should or should not show surface tension and sphericity, regardless of the detailed mechanism of biogenesis, are open question on which agnosticism is due [see Erdel and Rippe (2018); McSwiggen et al (2019a); McSwiggen et al (2019b); Peng and Weber, (2019) for further discussions]. It may be speculated that surface energy emerges from the density of the underlying mesh of interactions. Furthermore, we may surmise that macromolecules at the surface of a putative condensate have higher energy than those inside it. In this case, the surface energy may be expected to be high—and the boundary sharp—when the binding energies of the underlying molecular interactions are strong, which usually implies site specificity.
Most compartments may not result from PS
Thus, there are no good reasons to exclude SSIs as drivers of compartment assembly. Moreover, they are significantly more likely to participate in the biogenesis of the great diversity of subcellular compartments than a restricted variety of low‐affinity unspecific homotypic interactions. By way of example, the kinetochore is a compartment on chromosomes that concentrate at least 60 distinct polypeptides to control chromosome segregation (Musacchio & Desai, 2017). With the dissection of kinetochores well underway, all interactions characterized so far appear to be site specific, with discrete binding interfaces, whose mutation predictably prevents recruitment of downstream components. This does not reflect an absence of MVPs or IDPs, as several kinetochore components, including CENP‐CMif2 and KNL1Spc105, belong to these classes. In addition, we have no evidence that the kinetochore acts as an unspecific “sink” for functionally unrelated macromolecules. Its composition seems to be limited to functional components recruited to it in a site‐specific manner, as shown for BUB1/BUB3 and MAD2. This may be the norm, as the compositional identity of compartments is usually well defined. Readers are referred to a discussion on the centrosome as a condensate (Woodruff et al, 2017; Raff, 2019), and contextually to a report on how artificial coacervation promotes filamentation of a bacterial tubulin homolog (Te Brinke et al, 2018).
Seen retrospectively, it may seem puzzling that SSIs were excluded as an underlying possible driver of compartment assembly. Why were they? I suspect the answer is that the field, since early on and progressively more pervasively, accepted that compartments concentrate macromolecules through a mechanism of PS, and continued to support this idea, often in presence of very strong evidence to the contrary, without a rigorous test of the hypothesis, as I will show below. My concerns extend and complement previously formulated objections that many compartments indicated as condensates lack features expected if PS were their driver (Wheeler et al, 2016; McSwiggen et al, 2019b; Erdel et al, 2020). If many compartments do not show hallmarks of PS, and if their biogenesis is likely to reflect mechanisms other than those advocated by its proponents, we should avoid calling them condensates, which should be rather reserved for compartments with evident signs of condensation by PS.
Validation pipeline: part 1
So, there are compartments whose predominant and possibly only drivers of assembly are SSIs. Are there any compartments where PS is an undisputable driver of assembly? How should we investigate this question for other compartments where our knowledge of the assembly mechanisms is limited? As a thought experiment, let us consider a newly discovered membraneless compartment X with several concentrated macromolecules (Fig 5A). Measured FRAP rates and recovery fractions range from essentially no recovery for certain components to full recovery in minutes or seconds for others. These FRAP rates may be taken as an indication that condensate X is partly solid, partly liquid, but also as an indication that SSIs of variable strength are at stake. With hypotheses facing off, what should be done?
Figure 5. Interactions in an imaginary compartment.
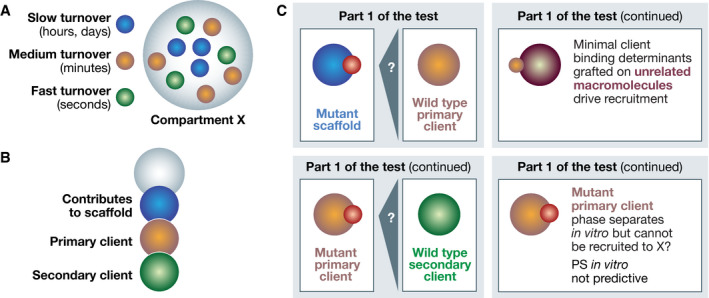
(A) Compartment X concentrates several components, three of which are shown in blue, brown, and green colors together with their turnover times; (B) An assembly hierarchy might have been identified while investigating the interactions in Compartment X; (C) Upper left: Can a primary client with wild‐type sequence be recruited to a scaffold with mutations in a binding site for the primary client? If so, the binding interaction is at least necessary for the recruitment. Upper right: Grafting of binding site of primary client for scaffold allows recruitment of unrelated macromolecule. Minimal binding sites indicate sufficiency. Bottom left: Iterative analysis down the interaction hierarchy. Bottom right: A mutant client that fails to be recruited to X but undergoes PS in vitro like the wild‐type counterpart indicates PS in vitro not predictive.
Using a combination of in vitro and in vivo work, SSIs supporters should aim to identify all components of compartment X and their binding regions, mutate them, and show which are crucial for assembly and what hierarchy exists, if any (Fig 5B). What should PS supporters do? Their hypothesis is that solvation exemplified by oil–water is crucial for compartment assembly, irrespective of any existing site‐specific interaction with other components. As it first needs to be proven that SSIs are at least insufficient for compartment assembly, one needs to identify, as a premise, all possible SSIs to assess how they contribute to assembly. This is precisely what those supposing SSIs planned to do, and that is how it should be: all possible hypotheses need to be considered, and SSIs should not be excluded from consideration without compelling reasons.
With the characterization of compartment X in progress, SSIs might have or have not been found. If they had not been found, compartment X might qualify for general PS. If they had been found, there would be a prospect for special PS but also for no PS at all. Thus, if SSIs have been found, the next question is whether they are necessary (and ideally sufficient) for compartment assembly in vivo. To assess this, a putative assembly scaffold may be mutated to eliminate the site‐specific binding determinant for a client (Fig 5C). In a strategy enabled by separation‐of‐function mutants, the mutant scaffold will assemble in its correct location if there remain binding sites for the compartment. If the client fails to become recruited, the SSI is at least necessary for recruitment in compartment X. This could be reapplied to the chain of primary, secondary, and higher‐order scaffold/clients. Under the PS paradigm, the continued recruitment of the mutant primary scaffold would imply that its solubility has not changed appreciably. If recruitment of the client, on the other hand, is abrogated, the PS model cannot invoke changes in its solubility in the bulk phase, as its sequence is invariant (the mutation is on the scaffold). Conversely, one may also mutate a client to prevent its recruitment into X, and ask if the mutation affects its PS behavior in vitro, i.e., whether PS in vitro is predictive of localization. In addition, as a criterion for sufficiency of SSIs, one may isolate the binding determinants of a client and graft them onto a macromolecule with entirely different features to ask if it becomes recruited in X, which would imply that the compartment does not discriminate for or against other features of the client (such as those that may determine its solubility).
Collectively, the pipeline described above is Part 1 of the test. With compartments being usually very complex, identifying interactions and assessing assembly hierarchies is an extremely demanding task requiring great biochemical prowess and a lot of patience, yielding modest short‐term rewards; but nonetheless essential to real progress.
Validation pipeline: part 2
With Part 1 complete, PS may be ready for limelight if SSIs in X were found to be completely absent, or if they were unnecessary or insufficient toward its biogenesis (in order of increasing likelihood). Part 2 of the test should aim to gather hard evidence for PS. The general PS idea implies that a condensate will appear when the concentration of a putative PS driver exceeds C sat (implying there should be a recognizable C sat). To test PS, we cannot manipulate the SSIs we might have identified in Part 1 of the test, as they are crucial thermodynamic parameters that have to remain unaltered when we are trying to assess additional effects of PS. Thus, we can only aim to change C sat, for instance, by increasing the relative concentration of candidate components of X in the dilute bulk phase so that they would not join X, either because they prefer it less or because they prefer the dilute phase more. But how is C sat determined in a cell? We have powerful theories for the behavior of associative polymers in “poor solvent” conditions (Hyman et al, 2014; Brangwynne et al, 2015; Harmon et al, 2017). But what about the cytosol, enormously complex, crowded, and offering a variety of unpredictable opportunities for competitive short‐lived unspecific interactions (Rivas & Minton, 2018)? The dense phase is also a complex compartment offering a variety of opportunities for short‐lived unspecific interactions. What determines the solubility in the dilute and condensed phases for any individual component? Is there a hard criterion for defining sufficiency? What exactly phase‐separates and defines the putative condensate? A single macromolecule? Its network of interactors? All components, as it seems plausible? How is a scaffold–client relationship maintained in this framework? How do post‐translational modifications influence this process? And are there additional energy‐consuming processes that further complicate the picture?
It is evident that if these questions were to be answered, Part 2 of the test would soon become an extremely complex endeavor, enormously more complex than the homotypic interaction assays that have been relied upon to prove general PS. In the majority of studies where general PS was claimed, SSIs were never considered as potential drivers of compartment biogenesis, even if clear evidence of their involvement already existed (two among many other cases are briefly discussed below). With Part 1 of the test generally skipped altogether, a poorly predictive and oversimplified version of Part 2 of the test made it all but impossible to fail. How is Part 2 of the test run in practice? Typically, it begins with the nomination of a plausible candidate for an associative polymer: MVPs, IDPs, or their fragments, but more recently essentially any macromolecular class. With experiments that sometimes demonstrate impressive technical and analytical depth, the putative PS driver is subjected to experiments in vitro aimed at deciphering its phase grammar and the rheological properties of the resulting droplets [e.g., Molliex et al (2015); Nott et al (2015); Pak et al (2016); Patel et al (2015); Wang et al (2018)]. If PS is observed and mutations that perturb it can be identified, which is almost invariably the case, the protein or its fragment is usually overexpressed in a cell together with mutants to build evidence that cellular behavior matches the results in vitro. In a majority of cases, the questions of how solubility of a putative PS scaffold is determined in vivo and whether a constant saturation concentration can be measured are not considered (see below).
Are general PS tests predictive?
This validation procedure, which has already led to the proposition of a large number of supposed PS “scaffolds,” has profound shortcomings (Protter et al, 2018). First, at sufficiently high concentrations, and often with help of crowding agents, there are virtually infinite conditions in vitro with the potential to modulate the solubility of a macromolecule until it crystallizes (rarely), or it precipitates or phase separates in droplets (less coveted, at least by crystallographers, but frequent) (Dumetz et al, 2008). These experiments can hardly be considered predictive of cellular behavior.
Second, it is unclear if the species that phase‐separates in vitro is a sufficiently faithful representation of the cellular counterpart. For example, divergence may result from incomplete modifications, proteolysis, conformational variability, or because the protein in vitro binds to a prominent contaminant, such as a chaperone. Verifying the precise chemical composition, conformation, and degree of homogeneity of a purported phase‐separated macromolecule in a cell is essentially unfeasible with our current means. Is the species in the compartment truly identical to its diluted counterpart? Is it homogenous? Is it precisely the same as it was tested in vitro?
Third, we should be wary of the way the scaffold–client conceptualization is molded to build a criterion for PS sufficiency. Many macromolecules identified as PS scaffolds are considered clients in the binding paradigm. For instance, NCK and N‐WASP are binding clients of phosphorylated nephrin (see below). Should these agents be sufficient for PS in cells as implied by their PS in vitro, they would assemble signaling compartments without activation cues, likely with catastrophic consequences.
Fourth, phase‐separated droplets in vitro may seem to mimic compartments observed in cells, but they only represent an initial, pre‐equilibrium stage of a process (Ostwald ripening, caused by minimization of the surface energy) that on a longer timescale promotes formation of a single macroscopic dense domain, an unappealing representation for most cellular compartments.
Fifth, how to interpret the outcome of PS experiments in vitro is uncertain and contentious. What significance should be attributed to (i) the formation of dense hydrogels, (ii) liquid droplets, (iii) cross‐β sheet interactions preluding to amyloid aggregation, or (iv) the various time‐dependent drop‐hardening phenomena [see, for instance, (Ader et al, 2010; Han et al, 2012; Kato et al, 2012, 2021; Burke et al, 2015; Kroschwald et al, 2015; Molliex et al, 2015; Nott et al, 2015; Patel et al, 2015; Xiang et al, 2015; Brady et al, 2017; Kato & McKnight, 2018; Martin & Mittag, 2018; Wang et al, 2018; Murthy et al, 2019; Martin et al, 2020; Borcherds et al, 2021)], for the actual structure and function of a condensate in vivo?
Finally, even if we had observed some correlation between the phase behavior of certain constructs in vitro and in vivo, we would remain entirely ignorant of how solubility of our putative driver is determined in the cell. There really is no good reason to expect that the solubility of putative PS drivers cherry‐picked for in vitro experiments matches their solubility in the cell. Additionally, the passive, thermodynamically driven experiment in vitro needs to confront the possibility of energy‐consuming, active processes present in (probably most) compartments, and possibly of stress responses caused by overexpression of constructs in the in vivo experiments.
Acceptance of complexity
Thus, the PS sufficiency tests that swaths of papers have employed to identify PS drivers greatly oversimplify the description of the mechanism of compartment assembly, and ought to have been given more credit for what they might imply than for what they effectively say. The infallibility of PS imputations based on these tests are a warning that validation standards lack robustness, but until recently these assays have been given full credit in major reviews (Brangwynne et al, 2015; Alberti et al, 2019). That these experiments are poorly predictive is exemplified by the complex effects of RNAs and other binding partners on PS of putative drivers, including FUS, TDP43, G3BP1/2, and many others [e.g., (Han et al, 2012; Schwartz et al, 2013; Burke et al, 2015; Kroschwald et al, 2015; Molliex et al, 2015; Patel et al, 2015; Zhang et al, 2015; Feric et al, 2016; Langdon et al, 2018; Maharana et al, 2018; Protter et al, 2018; Wang et al, 2018; Mann et al, 2019; Guillen‐Boixet et al, 2020; Riback et al, 2020; Sanders et al, 2020; Yang et al, 2020; Roden & Gladfelter, 2021)]. Only limitedly to FUS, for instance, its purported PS is inhibited by ATP, cell lysates, and RNA (Patel et al, 2017; Maharana et al, 2018; Protter et al, 2018). A possible suggestion to overcome the limits of the greatly error‐prone in vitro solubility tests for general PS currently in use is to run solubility experiments in cytomimetic media (Protter et al, 2018; Rivas & Minton, 2018, 2019; Gnutt et al, 2019; Nakashima et al, 2019). These should be realistically concentrated (100 mg/ml and more) and devoid of components of the putative condensate (else PS sufficiency cannot be demonstrated), e.g., bacterial cytosols or a realistic artificial medium built from the most dominant components of eukaryotic cytosols at their typical overall concentrations (Beck et al, 2011).
The multiphase dilution of PS
Recent work on the biogenesis of nuclear bodies, including the Cajal body and the nucleolus, and of other RNP compartments demonstrates that initial interpretations on the role of PS in their biogenesis were premature. The assembly of nuclear bodies mobilizes SSIs between various protein components and specific cis‐acting sequences on specific RNAs, and requires active transcription (Savino et al, 2001; Leung et al, 2004; Shav‐Tal et al, 2005; Kaiser et al, 2008; Shevtsov & Dundr, 2011; Grob et al, 2014; Berry et al, 2015; Caudron‐Herger et al, 2016; Falahati et al, 2016; Jain et al, 2016). Conversely, there does not seem to be strong biological evidence that these bodies originate from general PS. Nonetheless, this view has become dominant since it was shown that putative scaffolds in the nucleolus, nucleophosmin/NPM1, and fibrillarin, preferably in presence of RNA, phase‐separate in vitro to scaffold the assembly of immiscible phases reminiscent of the granular and dense fibrillar components, respectively (Brangwynne et al, 2011; Berry et al, 2015; Weber & Brangwynne, 2015; Feric et al, 2016). However suggestive, these observations need to be reconciled with a considerably more variegated picture. Nucleolar assembly is a complex regulated process involving several hundred components, spatial cues (e.g., the position of nucleolar organizer regions, or NORs, on chromosomes), and energy expenditures at various steps (Savino et al, 2001; Shav‐Tal et al, 2005; Boisvert et al, 2007, 2010). Recent super‐resolution fluorescence microscopy analyses of fibrillarin and other components of the dense fibrillar component, as well as of the rDNA and associated proteins, demonstrated a considerably more complex substructure than predicted by PS experiments in vitro (Yao et al, 2019; Maiser et al, 2020).
Recently, NPM1, shown to have a fixed C sat in isolation in vitro, was found not to have a fixed C sat when its concentration was progressively increased within intact cells (none of several additional putative PS drivers of other nuclear bodies had one either) (Riback et al, 2020). When mixed in vitro with rRNA and the nucleolar protein SURF6, again no fixed C sat was observed. Like many other nucleolar proteins, SURF6 contains a highly specific nucleolar localization signal (NoLS), known to bind NPM1 in multivalent configurations that can generate high binding affinities (Valdez et al, 1994; Kramer & Karpen, 1998; Mammen et al, 1998; Scott et al, 2010; Brabez et al, 2011; Mitrea et al, 2016). All evidence in vitro indicated that NPM1 and SURF6 bound each other, causing the ratio of NPM1 concentration in the dilute and dense phases to increase progressively as binding sites became saturated (Riback et al, 2020), a foregone expectation for a binding interaction.
In an undeterred new vision, the granular component of the nucleolus, in addition to the nucleolus itself, has been presented as a high‐dimensional condensate with multiple co‐influencing phases generated by heterotypic biomolecular interactions (Riback et al, 2020). Similar ideas were recently put forward for stress granules, another mainstay of general PS, after SSIs had re‐emerged there as the likely main drivers of assembly (Guillen‐Boixet et al, 2020; Sanders et al, 2020; Yang et al, 2020). While possibly formally correct, the description of compartments and parts thereof as high‐dimensional condensates is potentially deceiving: The new observations may not exclude PS, but do not implicate it either, while the complexity of this ad hoc description makes PS appear progressively less plausible. How would multidimensional condensates with hundreds of components with variable and co‐influencing C sat look like and function? This is precisely the point when the analogy loses appeal and must be replaced with real molecular understanding.
Special PS in the nuclear pore complex
Thus, general PS is a powerful analogy, but despite the flow of claims, evidence that it is the driver of biogenesis of any biological compartment of realistic complexity, a function for which it seems fundamentally inappropriate in comparison to SSIs, is thin, or probably even non‐existent. Better examples might eventually emerge, but will have to be scrutinized rigorously through shared criteria, with the established standards of research in molecular mechanistic biology that I presented above.
We should therefore discuss special PS, a “moderate” version of PS that assumes (i) that compartments are formed through SSIs, and (ii) that PS may manifest itself locally to meet a specific functional requirement. Although in vitro reconstitutions of phase‐separated compartments are also typical of studies of special PS, the motivation behind these experiments is different, as in special PS these experiments are not meant to identify putative drivers of compartment biogenesis. Rather, in special PS, these experiments are meant to test whether phase separation within the compartment can impart a specific functional feature. The best example involves the nuclear pore complex (NPC), a proteinaceous macromolecular complex that fuses the inner and outer nuclear membranes, generating a channel that crosses the nuclear envelope (Kabachinski & Schwartz, 2015; Hampoelz et al, 2019). The NPC assembles through SSIs of scaffold subunits, the scaffold nucleoporins (Scaffold‐Nups; Fig 6A), and appears as a complete membrane‐embedded ring with an inner cavity (Hampoelz et al, 2019).
Figure 6. PS in the cavity of the nuclear pore complex.
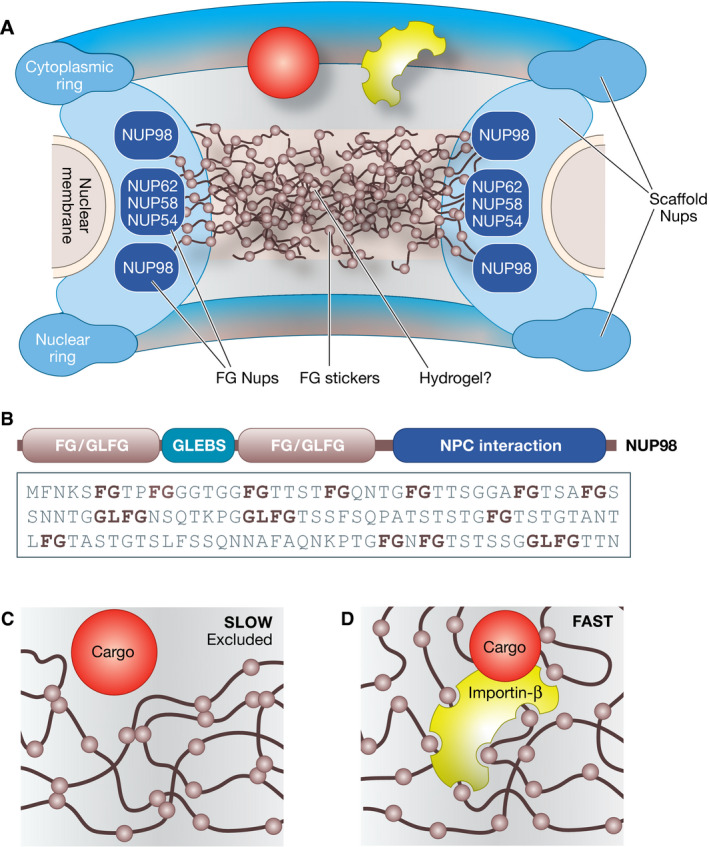
(A) The Scaffold‐Nups subunits of the NPC (not shown individually) build a complex circular structure with a central cavity. The FG‐Nups (only a subset of which are shown) interact with the Scaffold‐Nups through SSIs. This allows to position their IDRs in the cavity of the pore, where they may form a hydrogel; (B) Domain organization of human NUP98, one of the human FG‐Nups. A short segment of the FG/GLFG sequence is shown; (C) An inert cargo molecule of sufficiently large size will be unable to cross the NPC cavity; (D) The cargo needs to bind to an import/export receptor like importin‐β, which binds the FG repeats, locally “melting” the meshwork, and therefore dissolving in it.
The cavity is filled with the intrinsically disordered regions (IDRs) of a specialized subset of Nups, the FG‐Nups (Fig 6A). These contain large, low‐complexity disordered domains with characteristic phenylalanine glycine (FG) motifs and domains that bind the NPC scaffold via classical SSIs (Fig 6B) (Beck & Hurt, 2017). The FG‐Nups are required for the permeability barrier that controls material transfer between nucleoplasm and cytoplasm (Hurt, 1988; Wente et al, 1992; Patel et al, 2007; Hulsmann et al, 2012). Macromolecules with diameters above 4–5 nm or molecular masses above ~40 kDa fail to cross the NPC’s inner cavity with its ~50 nm diameter. Transport of these inert cargo macromolecules requires their interaction with a family of specialized nuclear transport receptors (NTRs) (Figs 1D and E, and 6C and D) (Bayliss et al, 1999; Gorlich & Kutay, 1999; Isgro & Schulten, 2005; Port et al, 2015). Thus, models of transport through the NPC need to explain what makes the majority of macromolecules inert and why the NTRs are selectively allowed to go through with their cargo (Frey et al, 2018).
The IDRs of FG‐Nups have an estimated concentration of 1 mM in the cavity of the NPC, and FG repeats may reach concentrations as high as 50 mM (Kabachinski & Schwartz, 2015; Schmidt & Gorlich, 2016; Beck & Hurt, 2017; Hampoelz et al, 2019). What material properties emerge from the clustering of the IDRs inside the NPC cavity? And how do these properties relate to the permeability barrier of the NPC? Several major competing hypotheses have emerged, covering the entire spectrum of possible phase behavior expected for IDRs (Lemke, 2016; Musser & Grunwald, 2016; Schmidt & Gorlich, 2016). In the virtual gate model, the IDRs of FG‐Nups, regarded as poorly cohesive, are proposed to align side by side, similarly to polyethylene glycol (PEG) molecules, to generate a “brush” that repels macromolecules but binds the NTRs, possibly coupled with a retraction mechanism upon NTR binding (Rout et al, 2003; Lim et al, 2007).
In the selective phase model (Ribbeck & Gorlich, 2001), on the other hand, the IDRs of a subset of cohesive FG‐Nups are proposed to undergo PS within the cavity of the NPC, driven by cohesive stacking and hydrophobic interactions of Phe residues in the FG repeats (Frey et al, 2006; Frey & Gorlich, 2007). Indeed, some of the most cohesive FG‐Nups stand out from a broader group, and more generally from IDPs, for a high level of hydrophobicity, comparable to that of folded globular proteins, and for almost negligible charge (no Arg, Lys, Asp, or Glu, and thus no repulsive forces) (Schmidt & Gorlich, 2015, 2016; Lemke, 2016). While PS of FG‐Nups does not occur outside the NPC without strong overexpression, FG hydrogels assemble in vitro from dilute solutions of FG‐Nups in aqueous buffers (Schmidt & Gorlich, 2015, 2016). Remarkably, these FG hydrogels recapitulate the main properties expected for the permselectivity barrier of the NPC, namely rejecting inert proteins of size larger than the hydrogel’s mesh size, and being a highly selective solvent for the NTRs (Frey et al, 2006; Frey & Gorlich, 2007).
The function of the NTRs is enabled by their being highly selective FG‐motif binders (Figs 1D and E, and 6C and D) (Iovine et al, 1995; Paschal & Gerace, 1995; Radu et al, 1995; Rexach & Blobel, 1995). Accordingly, NTRs are strikingly enriched on phenyl sepharose under highly stringent conditions (Ribbeck & Gorlich, 2002). Their navigation through the FG hydrogel is enabled by the ultrafast kinetics of their interactions with FG repeats, which involves multiple (probably up to eight or nine) FG‐binding sites on each NTR (Ribbeck & Gorlich, 2001; Isgro & Schulten, 2005; Frey & Gorlich, 2007; Milles et al, 2015; Port et al, 2015; Lemke, 2016). This allows NTRs and their associated cargo to cross the barrier within a few milliseconds (Yang et al, 2004), even if the overall binding affinity for FG motifs over multiple binding site on NTRs may be considerable. Thus, when the NTRs penetrate the mesh, they may temporarily dissolve the dense FG network (Schmidt & Gorlich, 2016).
The introduction of hexanediols, aliphatic alcohols, as tools to interfere with PS reflects the early ingenuous intuition that they might interfere specifically with the hydrophobic interactions between FG‐Nups and between FG‐Nups and NTRs, and thus interfere with hydrogel assembly and nuclear transport (Ribbeck & Gorlich, 2002). This approach had been developed in an age of innocence, but it is now becoming recognized that hexanediols have various negative consequences on cell physiology (Wheeler et al, 2016; Kroschwald et al, 2017; Duster et al, 2021; Itoh et al, 2021; Ulianov et al, 2021).
In the absence of direct ways to image the FG meshwork, biophysical work to test the selective phase hypothesis has been mainly performed in vitro. Work so far has been limited to especially cohesive FG‐Nups (Schmidt & Gorlich, 2015), thus falling short yet of explaining how the interplay of all FG‐Nups influences phase behavior and its consequences for transport through the NPC. Furthermore, FG‐Nups are strongly glycosylated, at least in metazoans (Labokha et al, 2013). How this modification influences cohesiveness and permselectivity, however, is unclear (Kabachinski & Schwartz, 2015; Schmidt & Gorlich, 2016). Finally, the interplay between FG‐Nups and NTRs remains poorly explored. These objections notwithstanding two decades of work on the selective phase hypothesis have generated a very large body of evidence in its support. Plausibly, it is the most advanced and best supported model of how phase properties of IDPs may cause PS and influence the material properties and function of a subcellular compartment. Anticipatory as it appears to be, this work has been puzzlingly ignored by the vast majority of reviews on the PS topic.
Frustrating complexity
The discussion of the selective phase hypothesis demonstrates that proving PS, even special PS, is enormously demanding. A discussion of multivalent systems further exemplifies this complexity. In Fig 7A and B, A and B are artificial MVPs containing multiple SH3 domains and PRMs (Li et al, 2012; Banani et al, 2016). In comparison to the interaction of individual SH3 and PRM modules (Fig 7A), the AB complex, with four interacting modules, can be expected to have very high affinity (Jencks, 1981; Mammen et al, 1998; Li et al, 2012). With concentrations of A and B increasing, the n stoichiometric AB complexes begin to interact in trans and cluster in distributed AnBn configurations (Li et al, 2012) (a simple A2B2 is shown in Fig 7B). This may drive PS (see below).
Figure 7. Complexity of a multivalent system.
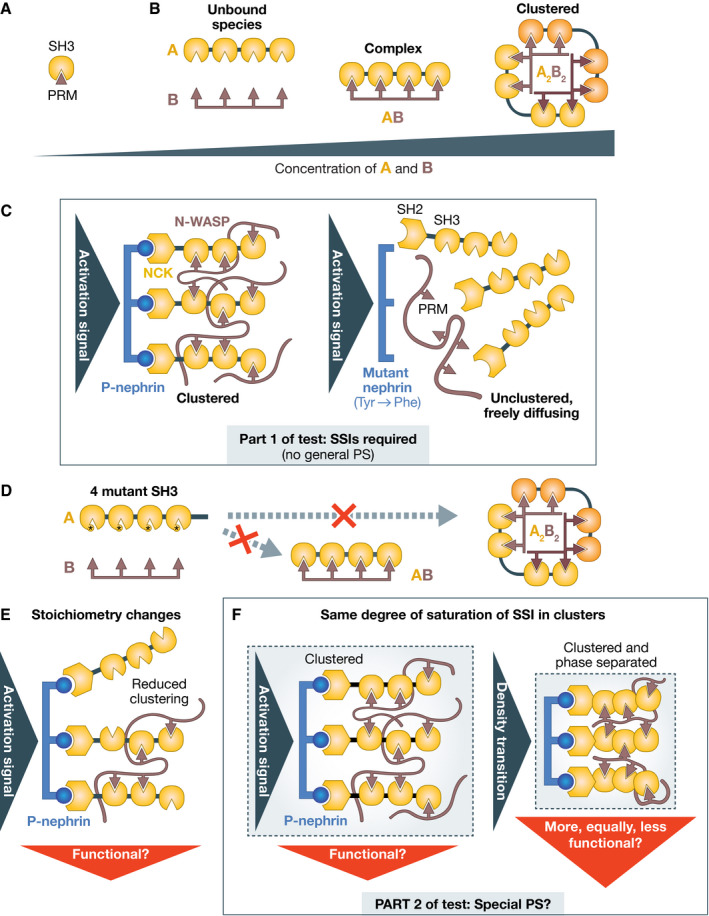
(A) A single SH3/proline‐rich motif module may have relatively low affinity; (B) Multiple modules on the two A and B ligands bind each other more tightly. The complex forms first as the concentration of A and B is increased. Further increasing the concentration leads to assemble clustered complexes in which multiple multivalent ligands interact at the same time in a network. At high concentrations, this system undergoes PS in vitro, demonstrating that PS is perfectly compatible with the SSIs that this system is based on. (C) The nephrin/NCK/N‐WASP system does not configure general PS because the SSIs elicited by phosphorylation of nephrin at three different sites are required for recruiting downstream components and membrane clustering. Clustering occurs when bound NCK promotes further recruitment of N‐WASP, which crosslinks the NCK molecules near P‐nephrin. Thus, binding interactions are required at all stages of assembly of this membrane‐bound signaling compartment; (D) Putative mutations at the SH3/PRM interface prevent formation of the simple AB complex as well as of the clustered A2B2 complex, indicating SSIs are necessary for both; (E) Changing stoichiometries in clustered compartments changes the degree of clustering, which may lead to graded functional responses. This strategy does not probe PS but rather binding saturation; (F) Part 2 of the test aims to detect PS within a compartment held together by SSIs. Ideally, PS should be measured under conditions of equal saturation of the binding interactions between all components. The solubility‐determining linkers between interacting “stickers” may determine whether a density transition has taken place or not. Functional output should be compared for the two depicted scenarios.
In the biological context that inspired the construction of A and B, nephrin, a transmembrane receptor activated by external ligands, causes phosphorylation‐dependent binding to the NCK SH2 domain and membrane localization of NCK and N‐WASP (broadly equivalent to A and B, respectively, as shown in Fig 1F). Nephrin is also multivalent (Fig 7C), as it displays three phosphorylation sites for NCK (Jones et al, 2006; Blasutig et al, 2008). It is an integral agent of the clustering mechanism shown schematically in Fig 7C (Case et al, 2019). NCK and N‐WASP are therefore binding clients of nephrin (Fig 3). After recruitment to nephrin, they turn into secondary binding scaffolds themselves and recruit proteins involved in actin polymerization (Ditlev et al, 2012; Su et al, 2016; Case et al, 2019). Thus, NCK and N‐WASP are not general PS scaffolds as the experiments with A and B might have seemed to imply (Li et al, 2012). If they were, their clustering would trigger (undesirable) signaling independently from upstream signaling events.
As shown in Fig 7C and D, mutations of the interfaces that mediate SSIs in the AB complex or in the nephrin/NCK/N‐WASP system would prevent all steps of complex assembly and clustering. PS in vitro in these systems (Li et al, 2012; Case et al, 2019) depends on the ability of the interacting regions of MVPs to form clustered arrangements. This demonstrates, beyond reasonable doubt and in addition to arguments that I raised previously, that SSIs are perfectly compatible with PS and liquid‐like behavior, at least in vitro. This mutational analysis, which corresponds to Part 1 of the test as defined above, also implies that this is not a case of general PS.
How could we test if this system undergoes special PS in vivo? A reasonable initial assumption is that the sparse distribution of phospho‐nephrin will drive formation of multivalent clusters of finite size, appearing as small dots in a fluorescence microscope (Case et al, 2019). In elegant experiments, it was shown that changes in the relative stoichiometries of the multivalent nephrin/NCK/N‐WASP ligands in membrane clusters affected downstream events, such as the rate of assembly of actin filaments by the Arp2/3 complex (Case et al, 2019). Changes in stoichiometry, however, may simply modulate the level of binding saturation of the interacting domains in the cluster, which in turn is expected to modulate downstream signaling events. They do not imply PS and in fact may appear to counter the idea of an underlying C sat (Fig 7E). Rather, because PS implies differential solvation and a density transition inside the compartment (Harmon et al, 2017), it should be tested under conditions in which the degree of saturation of binding interactions in the network remains approximately constant, as in the left and right panels of Fig 7F. Thus, despite a mention of PS in the title of the study (Case et al, 2019), whether this system undergoes PS in vivo is uncertain. Rather, we will need to distinguish between simple multivalent clusters and condensed, phase‐separated clusters (Fig 7F). How can this be approached?
In an initial theoretical treatment of PS in the simpler AB model system depicted in Fig 7B, PS was proposed to occur when the gain in configurational entropy compensates for the loss of translational and rotational entropy of the entities undergoing PS (Li et al, 2012). Later, this was partly amended and the increase in configurational entropy was rather proposed to drive clustering. In computational models, clustering might take the form of physical systems‐wide gelation (without PS) or of gelation driven by PS, the latter promoted by the specifics of the often long and disordered linkers between interacting domains (Banjade et al, 2015; Harmon et al, 2017). Even merely intuitively, focus on linkers is sensible because compartments lack long‐range order, and intervening flexible linkers allow domains to orient themselves freely for interactions with cognate modules. Conformational changes in the linkers may drive a more compact and dense organization.
In a real system like that in Fig 7F, clusters, as discussed above, are necessarily of finite size due to the overall small number of molecules available. Distinguishing between local gelation and local gelation driven by PS in these clusters will require manipulating the sequence of the interdomain linkers predicted to be the crucial solubility determinants (Banjade et al, 2015; Harmon et al, 2017; Choi et al, 2020), while measuring functional consequences of these manipulations ideally together with measurements of the density of the clusters (as it may be revealed by super‐resolution microscopy methods, for instance). However, in preparing for these experiments, there is a sobering gap between knowns and unknowns. We may have a theory for how the length and sequence of the interdomain linkers influence solubility in a defined buffer (Harmon et al, 2017; Choi et al, 2020), but we do not know how this will influence the interaction of the domain modules that the linkers separate. Model multivalent systems of identical motifs like AB may promote increases in configurational entropy more efficiently than systems made of more selectively interacting pairs, with a more restricted number of available configurations, the achievement of which may critically depend on linker length. Thus, besides solvation, linkers may also influence possible geometries restricting or favoring clustering. Linkers have rapidly diverging sequences, which complicates the formulation of simple, testable mechanistic hypotheses (van der Lee et al, 2014). Furthermore, we may not know many of the factors that determine solubility in the cellular environment, and should anticipate unexpected outcomes because of this. Thus, there are many thermodynamic variables in these experiments whose effects are difficult to control.
PS should explain something
Proving that PS exists and that it is functionally relevant, even in this second relatively simple system of purported special PS, will be an extremely difficult and tedious task. An examination of Fig 7F clarifies that there is no obvious reason why a simple cluster (left) should be any less functionally active than a phase‐separated cluster (right). In fact, the opposite may be true because a dense cluster may be expected to slow diffusion of effectors into the clusters. Thus, in this case, it is more difficult to grasp, in comparison to the NPC case, what functional need PS would address that could not be addressed without it.
The same concern can be expressed for the purported PS of the chromosomal passenger complex (CPC) (Trivedi et al, 2019). The CPC is a validated client of phosphorylation signals on centromeric chromatin (Fig 3) (Kelly et al, 2010; Wang et al, 2010; Yamagishi et al, 2010), but like various other examples, it was recast as an in vitro PS scaffold and proposed to phase‐separate after it reaches high concentrations at the centromere (Trivedi et al, 2019), with unclear functional gains. Neither of three related crucial tests in support of PS of the CPC was performed (Trivedi et al, 2019). First, it is unknown whether the cytosolic concentration of the CPC remains constant (C sat) during its accumulation at the centromere, a necessary condition to claim PS. Second, if PS was nucleated by concentration of the CPC at the centromere as proposed, PS should persist after removal of the initial spatial cues, in this case the phosphorylation signals that attract it to the centromere [see discussion in (Erdel & Rippe, 2018)]. Third, after nucleation of PS, additional CPC will be expected to be recruited independently of any spatial cues, including mutants unable to bind the phosphorylation cues [and independently of dimerization with the wild‐type complex (Bourhis et al, 2009; Bonner et al, 2020)].
Special PS may play a role in RNP granules (Protter et al, 2018), as well as on the surface of chromosomes, as suggested, for instance, by studies on Ki67, a surfactant that promotes the individualization of chromosomes during mitosis (Cuylen et al, 2016; Cuylen‐Haering et al, 2020). Chromosomes themselves, with a high local concentration of differently modified nucleosomes, may also be a favorable terrain for special PS. However, the technical and conceptual difficulties of studying special PS in this context are well exemplified by the controversy on the role of HP1 in the organization of heterochromatin (Larson et al, 2017; Strom et al, 2017; Sanulli et al, 2019; Erdel et al, 2020).
Conclusions
The study of PS in biology has grown explosively, sparking new and important research in polymer chemistry, material science, synthetic biology, origin of life, and the pathology of aggregation [e.g., (Jawerth et al, 2020; Reinkemeier & Lemke, 2021; Te Brinke et al, 2018; Wei et al, 2017)], often displaying an impressive degree of technical and intellectual sophistication. Part of the enormous success of the PS idea is driven by the legitimate anticipation that it will uncover new mechanisms in the area of neurodegenerative diseases and aggregative phenomena. Here, I exclusively focused on the molecular mechanisms of compartment assembly and on the potential role of PS in biology. I clarified that the apparent simplicity of the PS hypothesis is deceiving, as the general PS idea is based on rather implausible assumptions. Implausible does not imply erroneous. Nonetheless, the arguments presented here provide compelling evidence that the majority of PS claims have been based on highly incomplete tests that ignored more plausible drivers of macromolecular concentration. This criticism applies to the vast majority of PS claims in the literature, as can be easily verified retrospectively. Focusing on the mechanism of compartment assembly, my analysis echoes and complements a previous critique that questioned PS based on the appearance of compartments (McSwiggen et al, 2019b).
The study of the specificity of binding interactions builds on decades of tedious analyses that unveiled specificity rules and described them on the basis of their biochemistry, often providing very satisfactory explanations of the effects of mutation of (usually) conserved residues on phenotype. One should therefore provide very strong reasons in order to replace, or bypass, this framework with a new one, and any new framework should be expected to explain issues that were recalcitrant in the earlier view. Alternative hypotheses need to be considered and excluded (Part 1 and Part 2 of the test), but cannot be ignored. The colloidal science premises of the PS hypothesis were formulated in the first half of the twentieth century [see discussion in Hyman and Brangwynne (2011)], before the explosion of molecular biology, and ignorance of the crucial details made simplifications and analogies unavoidable back then. Nowadays, PS can only be a pillar of molecular mechanistic research.
The study of compartment biogenesis and organization should aim to reach the standards elucidated by work on the selective phase model (Ribbeck & Gorlich, 2001; Schmidt & Gorlich, 2016). The assembly principles of the NPC hydrogel are relatively simple in comparison to those of other cellular compartments. A major simplification is that NPC transport is thermodynamically driven, as it does not imply active processes within the NPC itself. Directionality of transport processes is ensured by gradients of the Ran GTPase outside the NPC (Schmidt & Gorlich, 2016). Compartments like the nucleolus show considerably greater complexity than the NPC. There is no simple pipeline to attack these structures and their (usually) non‐equilibrium dynamics. Modeling active processes in vitro with interacting parts capable of feedback control is a tremendous challenge that will project molecular mechanistic and synthetic biology into the future.
Even for simple compartments, there is more than a single laboratory can do, raising the question whether new models of cooperation are required. Progress will likely emerge from a combination of accurate part listings, complex reconstitutions, direct imaging (e.g., through cryo‐electron tomography and advanced super‐resolution fluorescence methods), simulation and theory, microfluidics, and more. The challenges ahead are substantial and so are the rewards, which include our future ability to understand and control biological objects and subcellular compartments of enormous complexity, including our chromosomes and the machinery that mediates intercellular communication.
Supporting information
Acknowledgements
I gratefully acknowledge financial support by the Max Planck Society, the European Research Council (ERC) SyG BIOMECANET (951430) and the Deutsche Forschung Gemeinschaft (DFG) Collaborative Research Centres (1093 and 1430). I am grateful to Simon Alberti, Amy Gladfelter, Tony Hyman, Tanja Mittag, Rohit Pappu, Michael Rosen, and Broder Schmidt for helpful discussions. I am very grateful to Susan Gasser, Roger Goody, Dirk Görlich, Stephen C. Harrison, Pim Huis in ‘t Veld, Roy Parker, Laurence Pearl, Thomas Surrey, Germán Rivas, Roland Winter, and Carl Wu for critical reading of the manuscript and for many helpful comments. Open Access funding enabled and organized by Projekt DEAL.
Disclosure statement and competing interests
The author declares that he has no conflict of interest.
The EMBO Journal (2022) 41: e109952.
See the Glossary for abbreviations used in this article.
References
- Ader C, Frey S, Maas W, Schmidt HB, Gorlich D, Baldus M (2010) Amyloid‐like interactions within nucleoporin FG hydrogels. Proc Natl Acad Sci USA 107: 6281–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Gladfelter A, Mittag T (2019) Considerations and challenges in studying liquid‐liquid phase separation and biomolecular condensates. Cell 176: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Hyman AA (2021) Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol 22: 196–213 [DOI] [PubMed] [Google Scholar]
- Aloy P, Ceulemans H, Stark A, Russell RB (2003) The relationship between sequence and interaction divergence in proteins. J Mol Biol 332: 989–998 [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK (2016) Compositional control of phase‐separated cellular bodies. Cell 166: 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjade S, Wu Q, Mittal A, Peeples WB, Pappu RV, Rosen MK (2015) Conserved interdomain linker promotes phase separation of the multivalent adaptor protein Nck. Proc Natl Acad Sci USA 112: E6426–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R, Ribbeck K, Akin D, Kent HM, Feldherr CM, Gorlich D, Stewart M (1999) Interaction between NTF2 and xFxFG‐containing nucleoporins is required to mediate nuclear import of RanGDP. J Mol Biol 293: 579–593 [DOI] [PubMed] [Google Scholar]
- Beck M, Hurt E (2017) The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18: 73–89 [DOI] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R (2011) The quantitative proteome of a human cell line. Mol Syst Biol 7: 549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP (2015) RNA transcription modulates phase transition‐driven nuclear body assembly. Proc Natl Acad Sci USA 112: E5237–5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasutig IM, New LA, Thanabalasuriar A, Dayarathna TK, Goudreault M, Quaggin SE, Li SS, Gruenheid S, Jones N, Pawson T (2008) Phosphorylated YDXV motifs and Nck SH2/SH3 adaptors act cooperatively to induce actin reorganization. Mol Cell Biol 28: 2035–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L et al (2018) Protein phase separation: a new phase in cell biology. Trends Cell Biol 28: 420–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Lam YW, Lamont D, Lamond AI (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics 9: 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner MK, Haase J, Saunders H, Gupta H, Li BI, Kelly AE (2020) The Borealin dimerization domain interacts with Sgo1 to drive Aurora B‐mediated spindle assembly. Mol Biol Cell 31: 2207–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherds W, Bremer A, Borgia MB, Mittag T (2021) How do intrinsically disordered protein regions encode a driving force for liquid‐liquid phase separation? Curr Opin Struct Biol 67: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis E, Lingel A, Phung Q, Fairbrother WJ, Cochran AG (2009) Phosphorylation of a borealin dimerization domain is required for proper chromosome segregation. Biochemistry 48: 6783–6793 [DOI] [PubMed] [Google Scholar]
- Brabez N, Lynch RM, Xu L, Gillies RJ, Chassaing G, Lavielle S, Hruby VJ (2011) Design, synthesis, and biological studies of efficient multivalent melanotropin ligands: tools toward melanoma diagnosis and treatment. J Med Chem 54: 7375–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady JP, Farber PJ, Sekhar A, Lin Y‐H, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman‐Kay JD et al (2017) Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell‐specific protein on phase separation. Proc Natl Acad Sci USA 114: E8194–E8203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324: 1729–1732 [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA (2011) Active liquid‐like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA 108: 4334–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Tompa P, Pappu RV (2015) Polymer physics of intracellular phase transitions. Nat Phys 11: 899–904 [Google Scholar]
- Te Brinke E, Groen J, Herrmann A, Heus HA, Rivas G, Spruijt E, Huck WTS (2018) Dissipative adaptation in driven self‐assembly leading to self‐dividing fibrils. Nat Nanotechnol 13: 849–855 [DOI] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL (2015) Residue‐by‐residue view of in vitro FUS granules that bind the C‐terminal domain of RNA polymerase II. Mol Cell 60: 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Zhang X, Ditlev JA, Rosen MK (2019) Stoichiometry controls activity of phase‐separated clusters of actin signaling proteins. Science 363: 1093–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron‐Herger M, Pankert T, Rippe K (2016) Regulation of nucleolus assembly by non‐coding RNA polymerase II transcripts. Nucleus 7: 308–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JM, Holehouse AS, Pappu RV (2020) Physical principles underlying the complex biology of intracellular phase transitions. Annu Rev Biophys 49: 107–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Yang JS, Choi Y, Ryu SH, Kim S (2009) Evolutionary conservation in multiple faces of protein interaction. Proteins 77: 14–25 [DOI] [PubMed] [Google Scholar]
- Courchaine EM, Lu A, Neugebauer KM (2016) Droplet organelles? EMBO J 35: 1603–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Blaukopf C, Politi AZ, Muller‐Reichert T, Neumann B, Poser I, Ellenberg J, Hyman AA, Gerlich DW (2016) Ki‐67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 535: 308–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen‐Haering S, Petrovic M, Hernandez‐Armendariz A, Schneider MWG, Samwer M, Blaukopf C, Holt LJ, Gerlich DW (2020) Chromosome clustering by Ki‐67 excludes cytoplasm during nuclear assembly. Nature 587: 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlev JA, Case LB, Rosen MK (2018) Who's in and Who's out‐compositional control of biomolecular condensates. J Mol Biol 430: 4666–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlev JA, Michalski PJ, Huber G, Rivera GM, Mohler WA, Loew LM, Mayer BJ (2012) Stoichiometry of Nck‐dependent actin polymerization in living cells. J Cell Biol 197: 643–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumetz AC, Chockla AM, Kaler EW, Lenhoff AM (2008) Protein phase behavior in aqueous solutions: crystallization, liquid‐liquid phase separation, gels, and aggregates. Biophys J 94: 570–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duster R, Kaltheuner IH, Schmitz M, Geyer M (2021) 1,6‐Hexanediol, commonly used to dissolve liquid‐liquid phase separated condensates, directly impairs kinase and phosphatase activities. J Biol Chem 296: 100260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum‐Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP (2015) The disordered P granule protein LAF‐1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USA 112: 7189–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rademacher A, Vlijm R, Tünnermann J, Frank L, Weinmann R, Schweigert E, Yserentant K, Hummert J, Bauer C et al (2020) Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1‐driven liquid‐liquid phase separation. Mol Cell 78: 236–249.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rippe K (2018) Formation of chromatin subcompartments by phase separation. Biophys J 114: 2262–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahati H, Pelham‐Webb B, Blythe S, Wieschaus E (2016) Nucleation by rRNA dictates the precision of nucleolus assembly. Curr Biol 26: 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fare CM, Villani A, Drake LE, Shorter J (2021) Higher‐order organization of biomolecular condensates. Open Biol 11: 210137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165: 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Gorlich D (2007) A saturated FG‐repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130: 512–523 [DOI] [PubMed] [Google Scholar]
- Frey S, Rees R, Schunemann J, Ng SC, Funfgeld K, Huyton T, Gorlich D (2018) Surface properties determining passage rates of proteins through nuclear pores. Cell 174: 202–217.e9 [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, Gorlich D (2006) FG‐rich repeats of nuclear pore proteins form a three‐dimensional meshwork with hydrogel‐like properties. Science 314: 815–817 [DOI] [PubMed] [Google Scholar]
- Gatlin JC, Matov A, Groen AC, Needleman DJ, Maresca TJ, Danuser G, Mitchison TJ, Salmon ED (2009) Spindle fusion requires dynein‐mediated sliding of oppositely oriented microtubules. Curr Biol 19: 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnutt D, Sistemich L, Ebbinghaus S (2019) Protein folding modulation in cells subject to differentiation and stress. Front Mol Biosci 6: 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SK, Mahamid J (2020) Visualizing molecular architectures of cellular condensates: hints of complex coacervation scenarios. Dev Cell 55: 97–107 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Grob A, Colleran C, McStay B (2014) Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev 28: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén‐Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlüßler R, Kim K, Trussina IREA, Wang J, Mateju D et al (2020) RNA‐induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181: 346–361.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampoelz B, Andres‐Pons A, Kastritis P, Beck M (2019) Structure and assembly of the nuclear pore complex. Annu Rev Biophys 48: 515–536 [DOI] [PubMed] [Google Scholar]
- Han T, Kato M, Xie S, Wu L, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G et al (2012) Cell‐free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149: 768–779 [DOI] [PubMed] [Google Scholar]
- Handwerger KE, Cordero JA, Gall JG (2005) Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low‐density, sponge‐like structure. Mol Biol Cell 16: 202–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon TS, Holehouse AS, Rosen MK, Pappu RV (2017) Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 6: e30294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED (2004) Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol 14: 953–964 [DOI] [PubMed] [Google Scholar]
- Hulsmann BB, Labokha AA, Gorlich D (2012) The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150: 738–751 [DOI] [PubMed] [Google Scholar]
- Hurt EC (1988) A novel nucleoskeletal‐like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae . EMBO J 7: 4323–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Brangwynne CP (2011) Beyond stereospecificity: liquids and mesoscale organization of cytoplasm. Dev Cell 21: 14–16 [DOI] [PubMed] [Google Scholar]
- Hyman AA, Simons K (2012) Cell biology. Beyond oil and water–phase transitions in cells. Science 337: 1047–1049 [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F (2014) Liquid‐liquid phase separation in biology. Annu Rev Cell Dev Biol 30: 39–58 [DOI] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR (1995) The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol 131: 1699–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isgro TA, Schulten K (2005) Binding dynamics of isolated nucleoporin repeat regions to importin‐beta. Structure 13: 1869–1879 [DOI] [PubMed] [Google Scholar]
- Itoh Y, Iida S, Tamura S, Nagashima R, Shiraki K, Goto T, Hibino K, Ide S, Maeshima K (2021) 1,6‐hexanediol rapidly immobilizes and condenses chromatin in living human cells. Life Sci Alliance 4: e202001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R (2016) ATPase‐modulated stress granules contain a diverse proteome and substructure. Cell 164: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, AlSadhan I, Vaidyanathan PP, Herschlag D (2020) How to measure and evaluate binding affinities. Elife 9: e57264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawerth L, Fischer‐Friedrich E, Saha S, Wang J, Franzmann T, Zhang X, Sachweh J, Ruer M, Ijavi M, Saha S et al (2020) Protein condensates as aging Maxwell fluids. Science 370: 1317–1323 [DOI] [PubMed] [Google Scholar]
- Jencks WP (1981) On the attribution and additivity of binding energies. Proc Natl Acad Sci USA 78: 4046–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ME, Hummer G (2011) Nonspecific binding limits the number of proteins in a cell and shapes their interaction networks. Proc Natl Acad Sci USA 108: 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N, Blasutig IM, Eremina V, Ruston JM, Bladt F, Li H, Huang H, Larose L, Li S‐C, Takano T et al (2006) Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature 440: 818–823 [DOI] [PubMed] [Google Scholar]
- Jones S, Thornton JM (1996) Principles of protein‐protein interactions. Proc Natl Acad Sci USA 93: 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabachinski G, Schwartz TU (2015) The nuclear pore complex–structure and function at a glance. J Cell Sci 128: 423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M (2008) De novo formation of a subnuclear body. Science 322: 1713–1717 [DOI] [PubMed] [Google Scholar]
- Kato M, Han T, Xie S, Shi K, Du X, Wu L, Mirzaei H, Goldsmith E, Longgood J, Pei J et al (2012) Cell‐free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, McKnight SL (2018) A solid‐state conceptualization of information transfer from gene to message to protein. Annu Rev Biochem 87: 351–390 [DOI] [PubMed] [Google Scholar]
- Kato M, Tu BP, McKnight SL (2021) Redox‐mediated regulation of low complexity domain self‐association. Curr Opin Genet Dev 67: 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H (2010) Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmazhan E, Tompa P, Dunn AR (2021) The role of ordered cooperative assembly in biomolecular condensates. Nat Rev Mol Cell Biol 22: 647–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RH, Karpen JW (1998) Spanning binding sites on allosteric proteins with polymer‐linked ligand dimers. Nature 395: 710–713 [DOI] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, Alberti S (2015) Promiscuous interactions and protein disaggregases determine the material state of stress‐inducible RNP granules. Elife 4: e06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Alberti S (2017) Hexanediol: a chemical probe to investigate the material properties of membrane‐less compartments. Matters. 10.19185/MATTERS.201702000010 [DOI] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hulsmann BB, Urlaub H, Baldus M, Gorlich D (2013) Systematic analysis of barrier‐forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J 32: 204–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM et al (2018) mRNA structure determines specificity of a polyQ‐driven phase separation. Science 360: 922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ (2017) Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547: 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke EA (2016) The multiple faces of disordered nucleoporins. J Mol Biol 428: 2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Gerlich D, Miller G, Lyon C, Lam YW, Lleres D, Daigle N, Zomerdijk J, Ellenberg J, Lamond AI (2004) Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J Cell Biol 166: 787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H‐C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF et al (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RY, Fahrenkrog B, Koser J, Schwarz‐Herion K, Deng J, Aebi U (2007) Nanomechanical basis of selective gating by the nuclear pore complex. Science 318: 640–643 [DOI] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R (2015) Formation and maturation of phase‐separated liquid droplets by RNA‐binding proteins. Mol Cell 60: 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon AS, Peeples WB, Rosen MK (2021) A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol 22: 215–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillén‐Boixet J, Franzmann TM et al (2018) RNA buffers the phase separation behavior of prion‐like RNA binding proteins. Science 360: 918–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiser A, Dillinger S, Langst G, Schermelleh L, Leonhardt H, Nemeth A (2020) Super‐resolution in situ analysis of active ribosomal DNA chromatin organization in the nucleolus. Sci Rep 10: 7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammen M, Choi SK, Whitesides GM (1998) Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed Engl 37: 2754–2794 [DOI] [PubMed] [Google Scholar]
- Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis‐Marks MR, Needham PG, Copley KE, Hurtle B, Portz B, Pyles NJ et al (2019) RNA binding antagonizes neurotoxic phase transitions of TDP‐43. Neuron 102: 321–338.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T (2020) Valence and patterning of aromatic residues determine the phase behavior of prion‐like domains. Science 367: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Mittag T (2018) Relationship of sequence and phase separation in protein low‐complexity regions. Biochemistry 57: 2478–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Hansen AS, Teves SS, Marie‐Nelly H, Hao Y, Heckert AB, Umemoto KK, Dugast‐Darzacq C, Tjian R, Darzacq X (2019a) Evidence for DNA‐mediated nuclear compartmentalization distinct from phase separation. Elife 8: e47098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Mir M, Darzacq X, Tjian R (2019b) Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev 33: 1619–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milles S, Mercadante D, Aramburu I, Jensen M, Banterle N, Koehler C, Tyagi S, Clarke J, Shammas S, Blackledge M et al (2015) Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell 163: 734–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW (2016) Nucleophosmin integrates within the nucleolus via multi‐modal interactions with proteins displaying R‐rich linear motifs and rRNA. Elife 5: e13571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy AC, Dignon GL, Kan Y, Zerze GH, Parekh SH, Mittal J, Fawzi NL (2019) Molecular interactions underlying liquid‐liquid phase separation of the FUS low‐complexity domain. Nat Struct Mol Biol 26: 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Desai A (2017) A molecular view of kinetochore assembly and function. Biology 6: 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser SM, Grunwald D (2016) Deciphering the structure and function of nuclear pores using single‐molecule fluorescence approaches. J Mol Biol 428: 2091–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima KK, Vibhute MA, Spruijt E (2019) Biomolecular chemistry in liquid phase separated compartments. Front Mol Biosci 6: 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott T, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett‐Jones D, Pawson T, Forman‐Kay J et al (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57: 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overlack K, Bange T, Weissmann F, Faesen AC, Maffini S, Primorac I, Muller F, Peters JM, Musacchio A (2017) BubR1 promotes Bub3‐Dependent APC/C inhibition during spindle assembly checkpoint signaling. Curr Biol 27: 2915–2927.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overlack K, Primorac I, Vleugel M, Krenn V, Maffini S, Hoffmann I, Kops GJ, Musacchio A (2015) A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. Elife 4: e05269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, Yunus AA, Liu DR, Pappu RV, Rosen MK (2016) Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol Cell 63: 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Gerace L (1995) Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol 129: 925–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee H, Jawerth L, Maharana S, Jahnel M, Hein M, Stoynov S, Mahamid J, Saha S, Franzmann T et al (2015) A liquid‐to‐solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162: 1066–1077 [DOI] [PubMed] [Google Scholar]
- Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, Hyman AA (2017) ATP as a biological hydrotrope. Science 356: 753–756 [DOI] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, Rexach MF (2007) Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129: 83–96 [DOI] [PubMed] [Google Scholar]
- Peng A, Weber SC (2019) Evidence for and against liquid‐liquid phase separation in the nucleus. Non‐coding RNA 5: 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peran I, Mittag T (2020) Molecular structure in biomolecular condensates. Curr Opin Struct Biol 60: 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T (2000) High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609 [DOI] [PubMed] [Google Scholar]
- Port SA, Monecke T, Dickmanns A, Spillner C, Hofele R, Urlaub H, Ficner R, Kehlenbach RH (2015) Structural and functional characterization of CRM1‐Nup214 interactions reveals multiple FG‐binding sites involved in nuclear export. Cell Rep 13: 690–702 [DOI] [PubMed] [Google Scholar]
- Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R (2018) Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep 22: 1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G (1995) The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 81: 215–222 [DOI] [PubMed] [Google Scholar]
- Raff JW (2019) Phase separation and the centrosome: a fait accompli? Trends Cell Biol 29: 612–622 [DOI] [PubMed] [Google Scholar]
- Reinkemeier CD, Lemke EA (2021) Dual film‐like organelles enable spatial separation of orthogonal eukaryotic translation. Cell 184: 4886–4903.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M, Blobel G (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83: 683–692 [DOI] [PubMed] [Google Scholar]
- Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei MT, Kriwacki RW, Brangwynne CP (2020) Composition‐dependent thermodynamics of intracellular phase separation. Nature 581: 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J 20: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D (2002) The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J 21: 2664–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas G, Minton AP (2018) Toward an understanding of biochemical equilibria within living cells. Biophys Rev 10: 241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas G, Minton AP (2019) Editorial: biochemical reactions in cytomimetic media. Front Mol Biosci 6: 145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden C, Gladfelter AS (2021) RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol 22: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Magnasco MO, Chait BT (2003) Virtual gating and nuclear transport: the hole picture. Trends Cell Biol 13: 622–628 [DOI] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A et al (2020) Competing protein‐RNA interaction networks control multiphase intracellular organization. Cell 181: 306–324.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ (2019) HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino TM, Gebrane‐Younes J, De Mey J, Sibarita JB, Hernandez‐Verdun D (2001) Nucleolar assembly of the rRNA processing machinery in living cells. J Cell Biol 153: 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Gorlich D (2015) Nup98 FG domains from diverse species spontaneously phase‐separate into particles with nuclear pore‐like permselectivity. Elife 4: e4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Gorlich D (2016) Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci 41: 46–61 [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Wang X, Podell ER, Cech TR (2013) RNA seeds higher‐order assembly of FUS protein. Cell Rep 5: 918–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ (2010) Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res 38: 7388–7399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW (2004) Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol 14: 942–952 [DOI] [PubMed] [Google Scholar]
- Shammas SL, Rogers JM, Hill SA, Clarke J (2012) Slow, reversible, coupled folding and binding of the spectrin tetramerization domain. Biophys J 103: 2203–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav‐Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D (2005) Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell 16: 2395–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M (2011) Nucleation of nuclear bodies by RNA. Nat Cell Biol 13: 167–173 [DOI] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382 [DOI] [PubMed] [Google Scholar]
- Snead WT, Gladfelter AS (2019) The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol Cell 76: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, McNally JG (2005) FRAP analysis of binding: proper and fitting. Trends Cell Biol 15: 84–91 [DOI] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH (2017) Phase separation drives heterochromatin domain formation. Nature 547: 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD (2016) Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352: 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudha G, Nussinov R, Srinivasan N (2014) An overview of recent advances in structural bioinformatics of protein‐protein interactions and a guide to their principles. Prog Biophys Mol Biol 116: 141–150 [DOI] [PubMed] [Google Scholar]
- Tanaka F (2011) Polymer physics: applications of molecular asssociation and thermoreversible gelation. Cambridge, UK: Cambridge University Press; [Google Scholar]
- Taylor NO, Wei MT, Stone HA, Brangwynne CP (2019) Quantifying dynamics in phase‐separated condensates using fluorescence recovery after photobleaching. Biophys J 117: 1285–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M (2008) Fuzzy complexes: polymorphism and structural disorder in protein‐protein interactions. Trends Biochem Sci 33: 2–8 [DOI] [PubMed] [Google Scholar]
- Trivedi P, Palomba F, Niedzialkowska E, Digman MA, Gratton E, Stukenberg PT (2019) The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat Cell Biol 21: 1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulianov SV, Velichko A, Magnitov MD, Luzhin A, Golov AK, Ovsyannikova N, Kireev II, Gavrikov A, Mishin A, Garaev AK et al (2021) Suppression of liquid‐liquid phase separation by 1,6‐hexanediol partially compromises the 3D genome organization in living cells. Nucleic Acids Res 49: 10524–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B (2015) Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett 589: 15–22 [DOI] [PubMed] [Google Scholar]
- Valdez BC, Perlaky L, Henning D, Saijo Y, Chan PK, Busch H (1994) Identification of the nuclear and nucleolar localization signals of the protein p120. Interaction with translocation protein B23. J Biol Chem 269: 23776–23783 [PubMed] [Google Scholar]
- van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT et al (2014) Classification of intrinsically disordered regions and proteins. Chem Rev 114: 6589–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink M, Simonetta M, Transidico P, Ferrari K, Mapelli M, De Antoni A, Massimiliano L, Ciliberto A, Faretta M, Salmon ED et al (2006) In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr Biol 16: 755–766 [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM (2010) Histone H3 Thr‐3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330: 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Choi J‐M, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D et al (2018) A molecular grammar governing the driving forces for phase separation of prion‐like RNA binding proteins. Cell 174: 688–699.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP (2015) Inverse size scaling of the nucleolus by a concentration‐dependent phase transition. Curr Biol 25: 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MT, Elbaum‐Garfinkle S, Holehouse AS, Chen CC, Feric M, Arnold CB, Priestley RD, Pappu RV, Brangwynne CP (2017) Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat Chem 9: 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP, Blobel G (1992) A new family of yeast nuclear pore complex proteins. J Cell Biol 119: 705–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R (2016) Distinct stages in stress granule assembly and disassembly. Elife 5: e18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA (2017) The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169: 1066–1077.e10 [DOI] [PubMed] [Google Scholar]
- Wu H, Fuxreiter M (2016) The structure and dynamics of higher‐order assemblies: amyloids, signalosomes, and granules. Cell 165: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL (2015) The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid‐like droplets, and nuclei. Cell 163: 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y (2010) Two histone marks establish the inner centromere and chromosome bi‐orientation. Science 330: 239–243 [DOI] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis R‐M, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q et al (2020) G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181: 325–345.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Gelles J, Musser SM (2004) Imaging of single‐molecule translocation through nuclear pore complexes. Proc Natl Acad Sci USA 101: 12887–12892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R‐W, Xu G, Wang Y, Shan L, Luan P‐F, Wang Y, Wu M, Yang L‐Z, Xing Y‐H, Yang LI et al (2019) Nascent pre‐rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol Cell 76: 767–783.e11 [DOI] [PubMed] [Google Scholar]
- Zhang H, Elbaum‐Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS (2015) RNA controls PolyQ protein phase transitions. Mol Cell 60: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Maslov S, Shakhnovich EI (2008) Constraints imposed by non‐functional protein‐protein interactions on gene expression and proteome size. Mol Syst Biol 4: 210 [DOI] [PMC free article] [PubMed] [Google Scholar]


