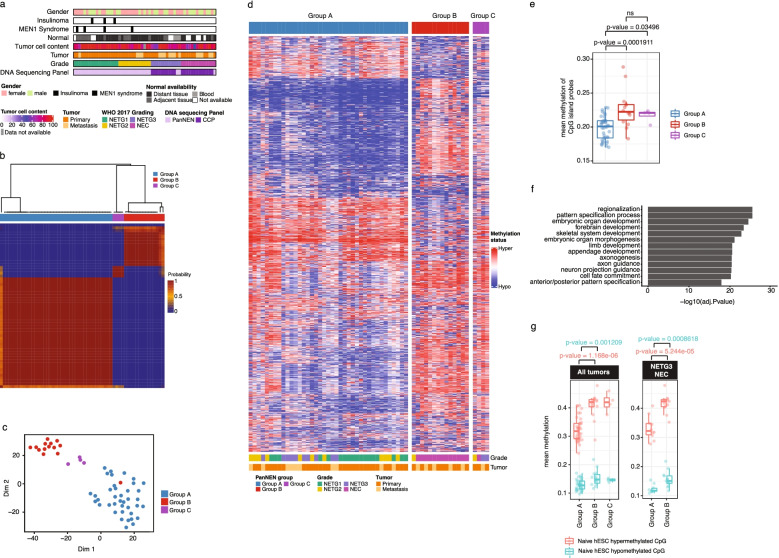
Fig. 1.
PanNENs subdivide into two main methylation groups. a Characterization of PanNEN cohort. WHO, World Health Organization; CCP, comprehensive cancer panel. b Unsupervised class discovery using 10,000 (10K) most variable methylation probes. Heatmap displays pairwise consensus values of the samples. c tSNE representation of PanNEN subgroups using 10K most variable probes. d. Heatmap displaying methylation status of 10K variable probes in each of the Groups A, B, and C. Methylation beta value was used to perform hierarchical clustering separately on each subgroup, identifying closely similar samples. Color range blue to red represents methylation beta value, columns indicate samples, and rows methylation probes. e Mean methylation of CpG island probes in PanNEN subgroups. Boxplot represents the distribution of mean methylation, each dot depicts a sample. f GO ontology analysis of 10K most variable probes, representing top 12 terms based on -Log10P-value. g Mean methylation of human Embryonic Stem Cells (hESC) associated hypermethylated and hypomethylated probes in PanNEN subgroups. Boxplot represents the distribution of hypermethylated (red) and hypomethylated (blue) CpG probes of hESC in cohort (left panel), or only in PanNETG3/ PanNEC samples from Group A and B respectively (right panel). Two-sample Wilcoxon test. Boxes show 25th and 75th percentiles and sample median as horizontal line, whiskers show maximum and minimum point