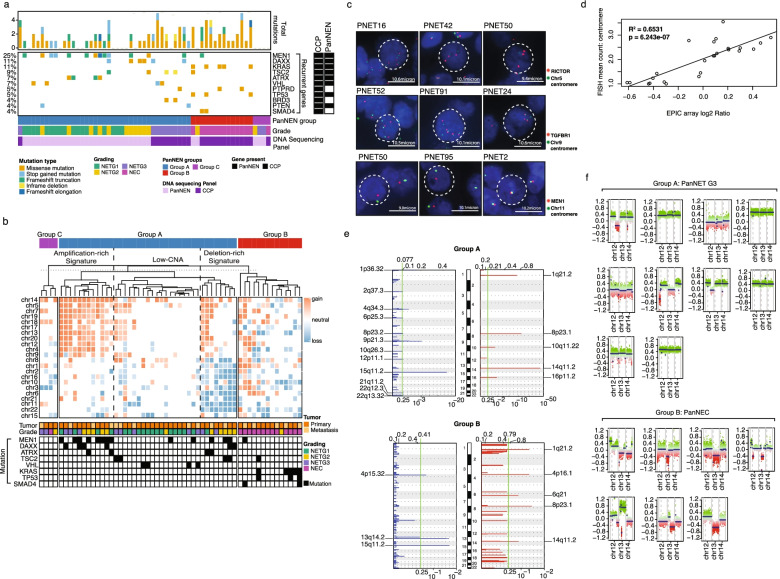
Fig. 2.
Genetic aberrations distinguish Group A and Group B. a Mutational landscape in PanNEN subgroups. Panel sequencing using in-house panel (PanNEN) and commercial cancer panel (CCP) Only genes mutated more than once are displayed here. Complete mutation profiles can be found in Additional file 2: Fig. S2b. Colors depict variant type (white spacing: no mutation identified). The DNA sequencing panel at the bottom depicts which targeted panel we used for the sample, while the ‘gene present’ annotation on the right side depicts whether the gene is present in the PanNEN panel or the CCP panel. Samples are sorted according to the PanNEN Groups; A, B and C. b Whole chromosomal aberrations in PanNEN subgroups. Hierarchical clustering of mean log2 ratios of chromosomal segments; dotted line represents cut-off used to identify amplification, low-CNA, and deletion-rich signatures; column annotation: tumor grade, tumor type and recurrently aberrated genes. c. Representative images of fluorescent in situ hybridization (FISH) validation. Red: gene probe. Green: centromere probe of chr5 (top panel), chr9 (middle panel), and chr11 (bottom panel). d Linear regression of mean copy number count of centromere derived from FISH (y-axis) and mean log2 ratios of chromosomal segments per autosome (x-axis); diagonal line: best fit model. R2 = 0.6531, p=6.232 × 10−7. e Focal aberrations in Group A (top panel) and Group B (bottom panel). Blue: focal copy number losses, red: focal copy number gain. Log2 ratio range at the top and q-value at the bottom of each graph. Green: q-value cut off at 0.25 to call significance. Significantly aberrated focal regions are identified. f Chromosome 12, 13, and 14 copy number status in NETG3 (top panel) and NEC (bottom panel). Intensity values of each bin are plotted in colored dots; each color indicates ‘methylated’ and ‘unmethylated’ channels of each CpG; segments are shown as horizontal blue lines