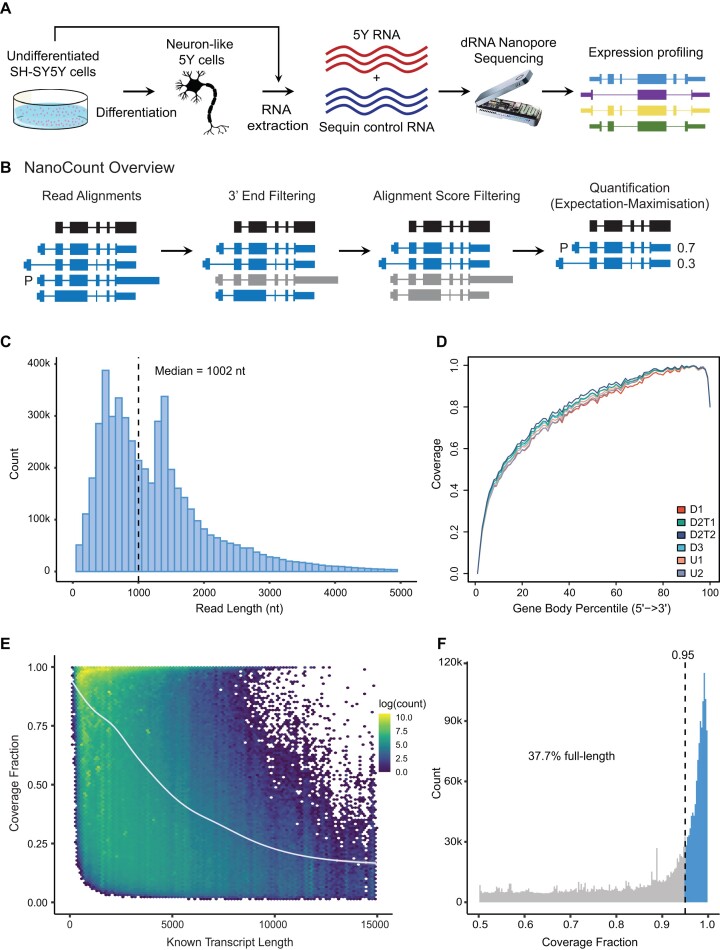
Figure 1.
Experimental overview and DRS read metrics. (A) Experimental overview. Cultured SH-SY5Y cells were differentiated in triplicate and RNA extracted from undifferentiated and differentiated cells. Native polyA purified SH-SY5Y RNA was combined with ‘sequin’ spike-in RNA, prepared for DRS and sequenced on an Oxford Nanopore MinION. Reads were analysed to identify and quantify genes and transcript isoforms and their differential expression. (B) Overview of NanoCount steps. A nanopore read (black) and example alignments (blue) are shown. P = primary alignment. Alignments with a 3′ end >50nt from the read 3′ end are discarded (grey). Next, alignments with an alignment score (AS) <95% of the highest remaining AS are discarded. Finally, the expectation-maximisation algorithm is initiated to quantify isoforms. In this example the read count is split 0.7 to 0.3 between the two remaining alignments. The primary alignment (P) is now the remaining alignment with the highest AS. (C) Length of all SH-SY5Y and sequin pass reads. Dashed line shows median read length. X-axis truncated at 5 kb. (D) Gene body coverage of SH-SY5Y reads in each sample. Length of all genes normalised to 100 and plotted from 5′ (0) to 3′ (100). Lines show mean coverage across all genes across the length of the gene body. Lower coverage at extreme 3' corresponds to soft clipping of the first bases sequenced which often have lower phred quality. U1 & U2—undifferentiated replicates 1 & 2. D1 & D3—differentiated replicates 1 & 3. D2T1 & T2—technical replicates of differentiated replicate sample 2. (E) Fraction of known transcript length covered by each read (coverage fraction) compared to known transcript length. Trend line was plotted using a generalized additive model, an extension of a generalised linear model where the linear form is replaced by sum of smooth functions. (F) Fraction of full-length SH-SY5Y reads. Fraction of known transcript length covered by each read. Dotted line represents 95% cutoff for full-length reads. Full-length reads shaded in blue. X-axis truncated at 0.5.