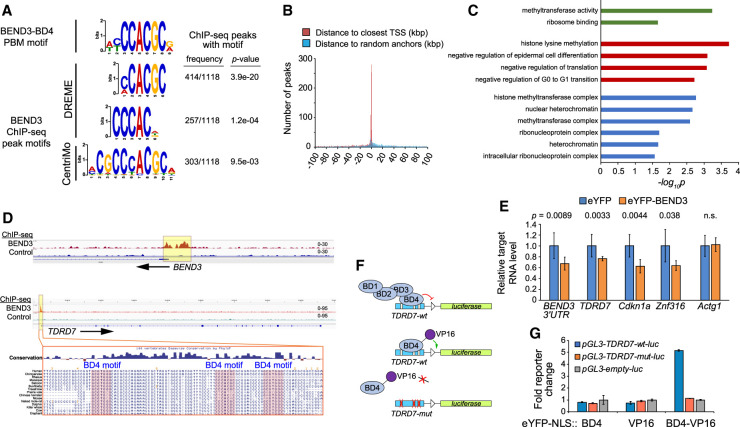
Figure 4.
BEND3 associates directly with BD4 motifs genome-wide. (A) The top motifs recovered from BEND3 ChIP-seq peaks from NTERA2 cells closely resemble the BEND3-BD4 motif identified from our in vitro binding assays. P-values of motif enrichment were generated using Fisher's exact test. (B) BEND3 peaks bearing BD4 sites are strongly localized near core promoters. (C) Representative gene ontology (GO) terms associated with BEND3 targets bearing BD4 sites. (D) IGV views of BEND3 and control ChIP-seq data at BEND3 and TDRD7. BEND3 ChIP-seq peaks in both genes are associated with multiple BD4 sites, as illustrated for TDRD7. (E) Transcript levels of BEND3-BD4 target genes are repressed upon ectopic expression of eYFP-BEND3. (F) Schematic of TDRD7-wt and mutant transcriptional reporters, and rationale to infer direct recruitment of BD4 to its cognate sites in TDRD7 using a BD4-VP16 activator fusion. (G) Luciferase reporter assays show that BD4 and VP16 domains are neutral, but BD4-VP16 specifically activates the TDRD7-wt reporter. Data in E and G are mean ± SD; n = 3 experiments; two-tailed t-test applied.