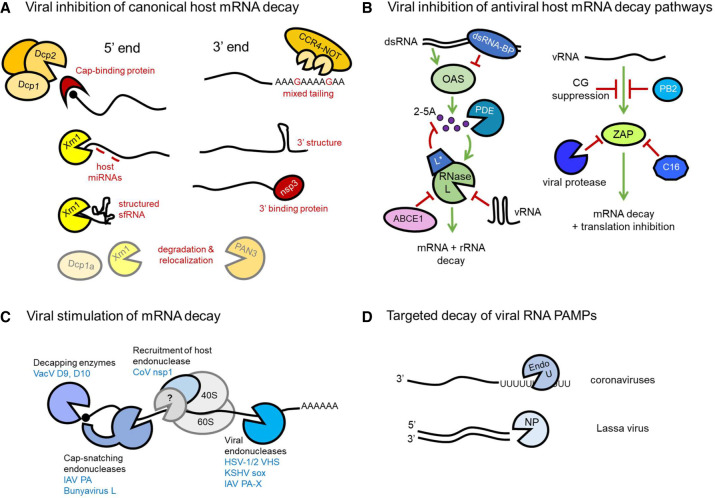
Figure 1.
Viral strategies to oppose and promote mRNA decay. (A) Inhibition of cellular proteins that effect 5′–3′ mRNA decay can be achieved by protection from decapping by cap binding proteins (VPg) and obstruction of the exonuclease Xrn1 by miRNAs and RNA structure (sfRNA). 3′ end decay is similarly opposed by recruitment of viral proteins and formation of RNA structure, and “mixed tailing” can also inhibit poly(A) tail deadenylation by CCR4–NOT. Both 5′ and 3′ targeting cellular mRNA decay proteins may be degraded or relocalized during infection. (B) Viruses inhibit OAS/RNase L and ZAP antiviral cellular RNA decay pathways using diverse strategies. Recognition of dsRNA by OAS is blocked by viral dsRNA-BPs. Viral phosphodiesterases (PDEs) degrade second messenger 2-5A. RNase L activation can also be blocked by viral proteins, up-regulated cellular negative regulator ABCE1, or viral RNA structures. ZAP recognition of virus RNA (vRNA) is evaded by CG suppression, viral ZAP-binding proteins, and ZAP cleavage by virus proteases. (C) Viral proteins stimulate mRNA decay via multiple modalities. They can contain direct enzymatic activity such as mRNA decapping and endonucleolytic cleavage, which may be cap-proximal. In contrast, coronavirus nsp1 binds the 40S ribosome and, although no direct nucleolytic activity has been identified, effects mRNA cleavage in infected cells, leading to speculation that it recruits a host decay enzyme. (D) Viral RNA PAMPs are directly controlled by viral dsRNA-specific exonucleases or U-specific endonucleases that, by limiting the potential for 5′ (−)-strand poly(U) sequences to complex with A-rich sequences, prevent dsRNA formation.