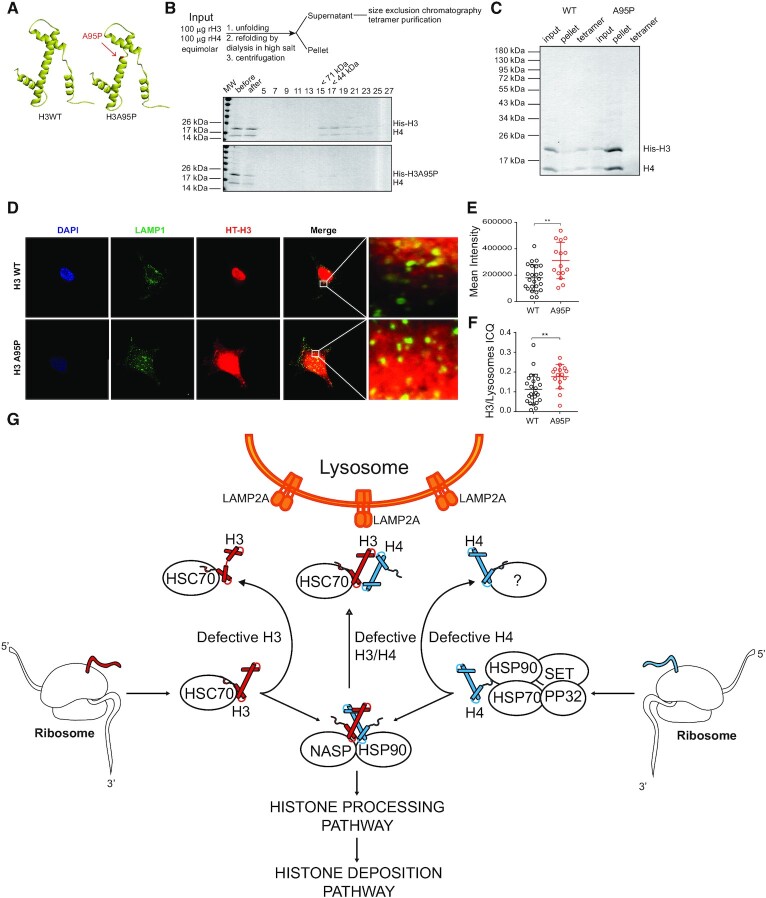
Figure 6.
CMA participates in the degradation of misfolded histone H3 protein. (A) Ribbon diagram illustrating the structures of wild-type histone H3 and mutant histone H3A95P. The arrow points to the site where A95 is located along the second α-helix and the disruption of the helix upon mutation to proline. (B) Top, scheme illustrating the experimental procedure to assemble tetramers. Bottom, Coomassie blue staining analysis of fractions derived from the S200 sizing column after tetramer formation utilizing wild type His-tagged or His-H3A95P. (C) Coomassie blue staining gel of samples derived from the tetramer assembly reaction (input, pellet and tetramer derived from the S200 column) with wild type His-tagged and His-H3A95P. (D–F) Immunofluorescence of HeLa cells for LAMP1 (green), DAPI (blue), and HT-H3 or -H3A95P (red) (D). Mean intensity in arbitrary units of HT-H3 signal of 20 cells per condition (E). Intensity Correlation Quotient (ICQ) graph, as HT/LAMP1 signal of 20 cells per condition (F); **P< 0.01, Student′s t-test. (G) Model for the quality control of newly synthesized histones H3 and H4. The figure illustrates the relation between the biogenesis and the degradation mediated by Chaperone Mediated Autophagy (CMA) of histones H3 and H4 in the cytoplasm. As shown in this work, CMA participates in the degradation of histones H3 and H4 and possibly in the quality control of histones by degrading unfolded or defective newly synthesized histones H3 and H4. The chaperones HSC70 and tNASP are important regulators in this pathway, in such a way that HSC70 can either assist in the proper folding of histones or deliver H3 to the LAMP2A receptor at the lysosome for degradation. Whether histone H4 is also targeted by HSC70 to the lysosome is currently unknown. tNASP instead stabilizes and prevents histones H3 and H4 degradation.